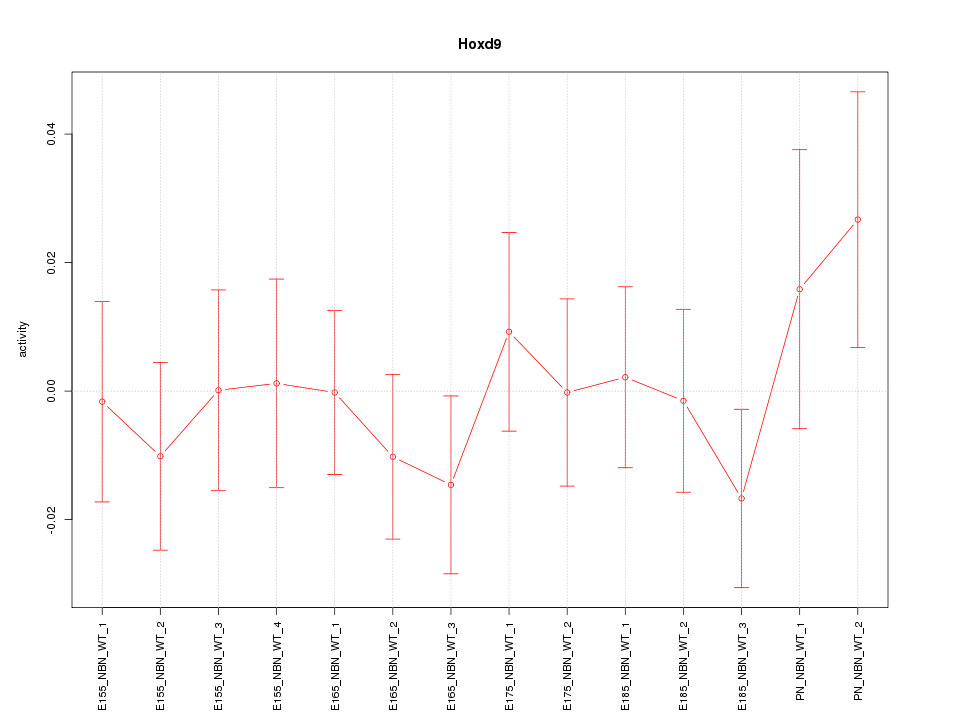
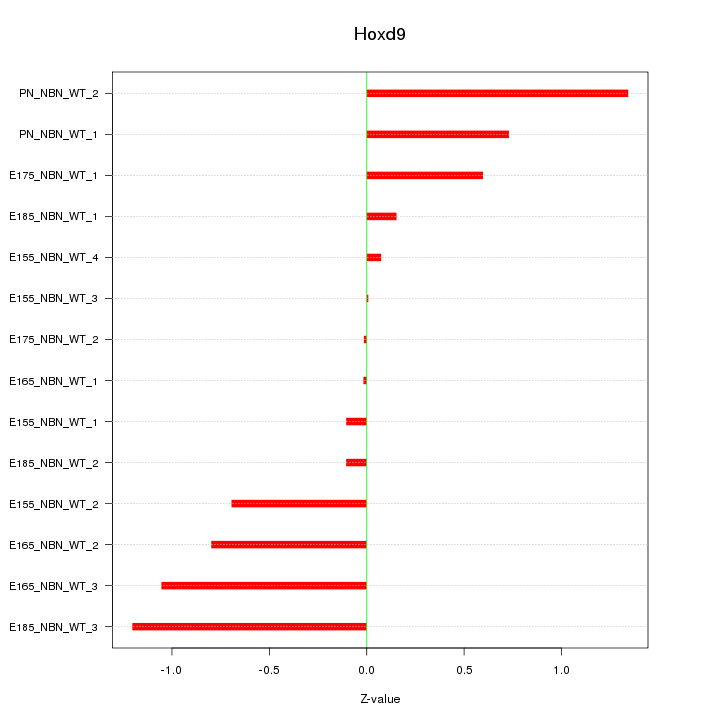
Motif ID: Hoxd9
Z-value: 0.677
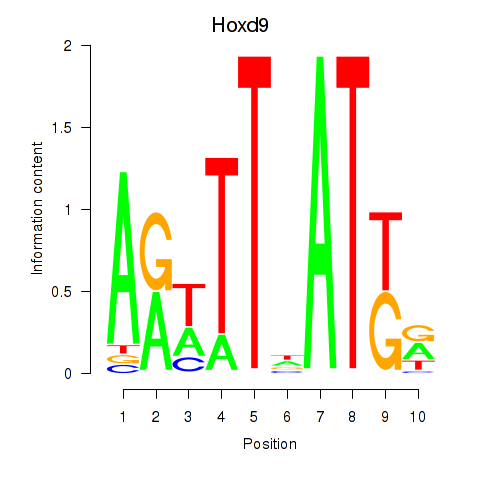
Transcription factors associated with Hoxd9:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hoxd9 | ENSMUSG00000043342.8 | Hoxd9 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.3 | 1.0 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 0.3 | 1.0 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.2 | 1.5 | GO:0035989 | tendon development(GO:0035989) |
| 0.2 | 0.9 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.4 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.1 | 0.4 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.1 | 0.7 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.1 | 0.5 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.1 | 0.8 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.1 | 0.3 | GO:1904395 | regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.1 | 0.4 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.1 | 0.4 | GO:0098915 | membrane repolarization during ventricular cardiac muscle cell action potential(GO:0098915) |
| 0.1 | 0.5 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.1 | 0.3 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.1 | 1.9 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 1.0 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 0.4 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.4 | GO:1901409 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 1.5 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.1 | GO:2000569 | T-helper 2 cell activation(GO:0035712) positive regulation of T-helper 17 type immune response(GO:2000318) regulation of T-helper 2 cell activation(GO:2000569) positive regulation of T-helper 2 cell activation(GO:2000570) |
| 0.0 | 0.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.5 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 | 0.4 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.0 | 0.8 | GO:0032148 | activation of protein kinase B activity(GO:0032148) |
| 0.0 | 0.2 | GO:0015840 | urea transport(GO:0015840) urea transmembrane transport(GO:0071918) |
| 0.0 | 0.5 | GO:0071398 | cellular response to fatty acid(GO:0071398) |
| 0.0 | 0.2 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.0 | 0.4 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.1 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.0 | 1.3 | GO:0050810 | regulation of steroid biosynthetic process(GO:0050810) |
| 0.0 | 0.1 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.0 | 0.5 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.1 | GO:1903056 | regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.0 | 0.8 | GO:0008542 | visual learning(GO:0008542) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 1.5 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.4 | GO:0071438 | NADPH oxidase complex(GO:0043020) invadopodium membrane(GO:0071438) |
| 0.1 | 0.3 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.1 | 1.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 1.2 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 1.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 1.0 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.3 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 0.4 | GO:0005911 | cell-cell junction(GO:0005911) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.2 | 1.5 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 0.5 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.1 | 0.4 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 0.4 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.1 | 0.5 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.1 | 0.5 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.1 | 0.3 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.1 | 1.0 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.1 | 0.4 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 1.2 | GO:0017166 | dystroglycan binding(GO:0002162) vinculin binding(GO:0017166) |
| 0.1 | 1.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 1.0 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.1 | 0.4 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 0.7 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 1.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.8 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.0 | 0.8 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 1.0 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.5 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |