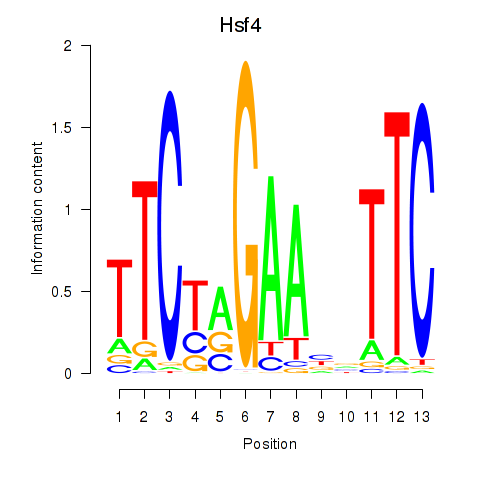
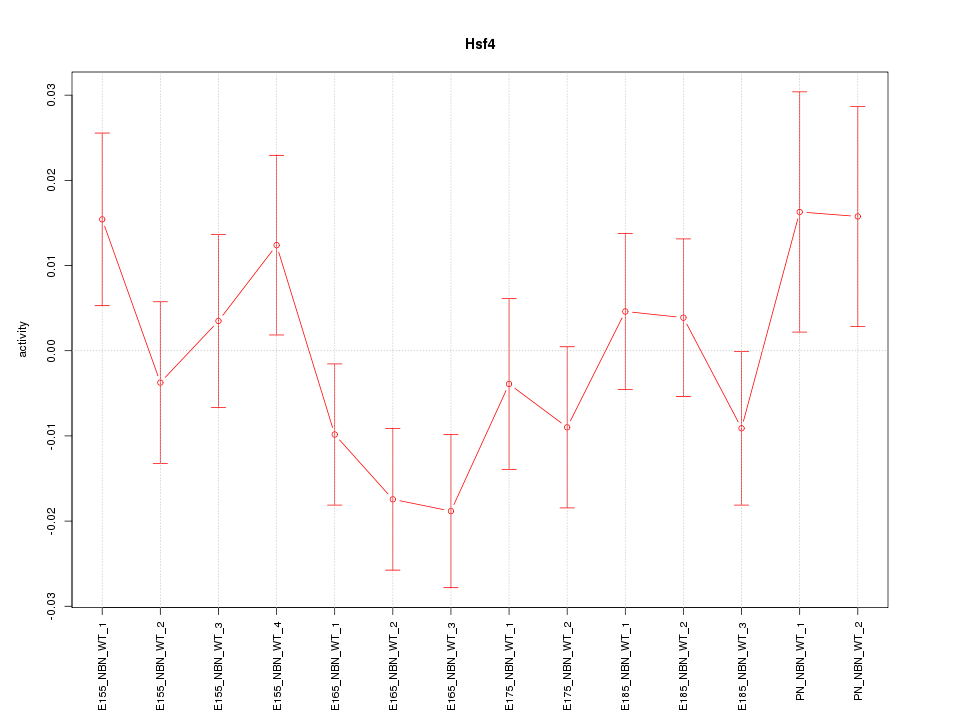
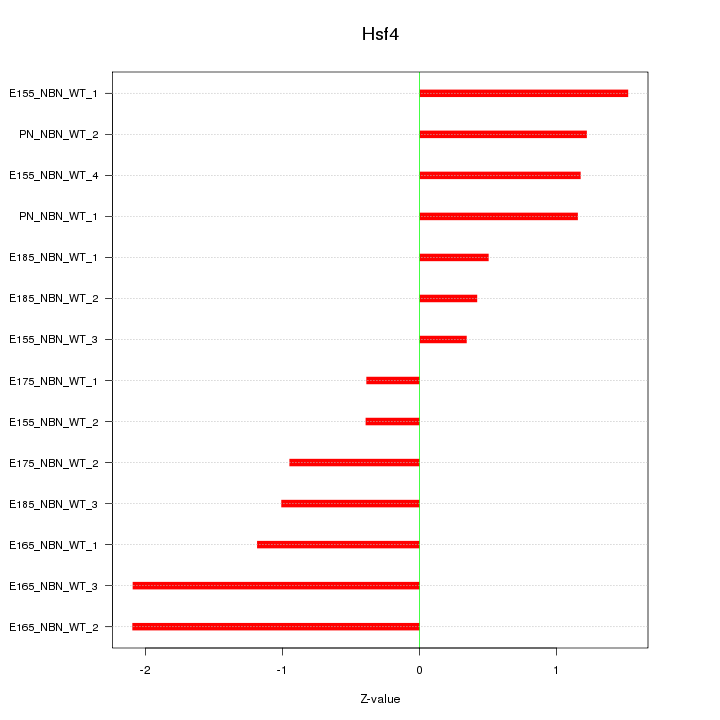
| 0.6 |
3.3 |
GO:0045583 |
regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.5 |
1.4 |
GO:1900041 |
negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.5 |
1.9 |
GO:0042938 |
dipeptide transport(GO:0042938) |
| 0.5 |
1.4 |
GO:0035799 |
ureter maturation(GO:0035799) |
| 0.4 |
1.4 |
GO:0044565 |
dendritic cell proliferation(GO:0044565) |
| 0.3 |
1.0 |
GO:0033364 |
mast cell secretory granule organization(GO:0033364) |
| 0.3 |
1.0 |
GO:0035701 |
hematopoietic stem cell migration(GO:0035701) |
| 0.3 |
1.5 |
GO:1905049 |
negative regulation of metalloendopeptidase activity(GO:1904684) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.3 |
0.8 |
GO:0010845 |
positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.3 |
1.3 |
GO:0034421 |
post-translational protein acetylation(GO:0034421) |
| 0.2 |
1.0 |
GO:0002457 |
T cell antigen processing and presentation(GO:0002457) |
| 0.2 |
0.6 |
GO:0031117 |
serotonin receptor signaling pathway(GO:0007210) positive regulation of microtubule depolymerization(GO:0031117) |
| 0.2 |
0.5 |
GO:0090187 |
positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.2 |
1.3 |
GO:0017196 |
N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.2 |
0.5 |
GO:0001732 |
formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.2 |
1.6 |
GO:0032876 |
negative regulation of DNA endoreduplication(GO:0032876) |
| 0.2 |
0.5 |
GO:0002295 |
T-helper cell lineage commitment(GO:0002295) interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.2 |
0.6 |
GO:0060715 |
syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.1 |
0.7 |
GO:0031033 |
myosin filament organization(GO:0031033) |
| 0.1 |
0.6 |
GO:0035063 |
nuclear speck organization(GO:0035063) |
| 0.1 |
0.7 |
GO:0006564 |
L-serine biosynthetic process(GO:0006564) |
| 0.1 |
1.7 |
GO:0061299 |
retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 |
0.6 |
GO:0006369 |
termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 |
0.4 |
GO:0021529 |
spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.1 |
0.4 |
GO:0008228 |
opsonization(GO:0008228) |
| 0.1 |
1.0 |
GO:0090370 |
negative regulation of cholesterol efflux(GO:0090370) |
| 0.1 |
1.0 |
GO:0006620 |
posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 |
0.3 |
GO:0007228 |
positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.1 |
0.8 |
GO:0006027 |
glycosaminoglycan catabolic process(GO:0006027) |
| 0.1 |
0.6 |
GO:0002553 |
histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.1 |
0.6 |
GO:0048254 |
snoRNA localization(GO:0048254) |
| 0.1 |
0.5 |
GO:0051012 |
microtubule sliding(GO:0051012) |
| 0.1 |
0.6 |
GO:0070447 |
positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.1 |
0.9 |
GO:0035881 |
amacrine cell differentiation(GO:0035881) |
| 0.1 |
1.1 |
GO:0031936 |
negative regulation of chromatin silencing(GO:0031936) |
| 0.1 |
0.4 |
GO:0002924 |
negative regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002924) negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.1 |
0.7 |
GO:1902018 |
negative regulation of cilium assembly(GO:1902018) |
| 0.1 |
0.6 |
GO:0006590 |
thyroid hormone generation(GO:0006590) |
| 0.1 |
0.5 |
GO:0007021 |
tubulin complex assembly(GO:0007021) |
| 0.1 |
0.5 |
GO:0060178 |
regulation of exocyst localization(GO:0060178) |
| 0.1 |
0.3 |
GO:0001803 |
antibody-dependent cellular cytotoxicity(GO:0001788) type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 0.1 |
0.3 |
GO:0009414 |
response to water deprivation(GO:0009414) |
| 0.1 |
3.0 |
GO:0002474 |
antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 |
0.6 |
GO:0019348 |
dolichol metabolic process(GO:0019348) |
| 0.1 |
0.2 |
GO:0019043 |
establishment of viral latency(GO:0019043) |
| 0.1 |
0.3 |
GO:0042535 |
positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.1 |
0.5 |
GO:0000056 |
ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 |
0.3 |
GO:0034145 |
positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.1 |
0.2 |
GO:0032918 |
polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 |
0.6 |
GO:0034498 |
early endosome to Golgi transport(GO:0034498) |
| 0.1 |
0.3 |
GO:2000346 |
negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 |
0.8 |
GO:0019262 |
N-acetylneuraminate catabolic process(GO:0019262) |
| 0.1 |
0.7 |
GO:0090051 |
negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.1 |
0.2 |
GO:0006556 |
S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 |
0.2 |
GO:0042228 |
interleukin-8 biosynthetic process(GO:0042228) |
| 0.1 |
0.2 |
GO:0071492 |
cellular response to UV-A(GO:0071492) |
| 0.1 |
0.7 |
GO:0000042 |
protein targeting to Golgi(GO:0000042) |
| 0.1 |
0.3 |
GO:0000098 |
sulfur amino acid catabolic process(GO:0000098) |
| 0.1 |
0.8 |
GO:0070986 |
left/right axis specification(GO:0070986) |
| 0.1 |
0.2 |
GO:0007056 |
spindle assembly involved in female meiosis(GO:0007056) positive regulation of oocyte development(GO:0060282) |
| 0.1 |
0.5 |
GO:0042359 |
vitamin D metabolic process(GO:0042359) |
| 0.1 |
0.1 |
GO:2000298 |
regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 |
0.4 |
GO:0090037 |
positive regulation of protein kinase C signaling(GO:0090037) |
| 0.0 |
0.1 |
GO:1902524 |
negative regulation of interferon-alpha production(GO:0032687) interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) negative regulation of interferon-beta biosynthetic process(GO:0045358) positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.0 |
0.7 |
GO:0016926 |
protein desumoylation(GO:0016926) |
| 0.0 |
0.2 |
GO:1901252 |
regulation of intracellular transport of viral material(GO:1901252) |
| 0.0 |
0.3 |
GO:0090383 |
phagosome acidification(GO:0090383) |
| 0.0 |
0.1 |
GO:1904154 |
protein localization to endoplasmic reticulum exit site(GO:0070973) positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 |
0.2 |
GO:0002326 |
B cell lineage commitment(GO:0002326) |
| 0.0 |
0.1 |
GO:0090526 |
glucosamine metabolic process(GO:0006041) regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 |
0.2 |
GO:2000048 |
negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 |
0.6 |
GO:0060052 |
neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 |
0.1 |
GO:0009838 |
abscission(GO:0009838) |
| 0.0 |
2.4 |
GO:0006749 |
glutathione metabolic process(GO:0006749) |
| 0.0 |
0.2 |
GO:0045650 |
negative regulation of macrophage differentiation(GO:0045650) |
| 0.0 |
0.5 |
GO:0035988 |
chondrocyte proliferation(GO:0035988) |
| 0.0 |
0.3 |
GO:0006729 |
tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 |
0.3 |
GO:0045779 |
negative regulation of bone resorption(GO:0045779) |
| 0.0 |
0.7 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 |
0.3 |
GO:0030575 |
nuclear body organization(GO:0030575) |
| 0.0 |
1.1 |
GO:0045494 |
photoreceptor cell maintenance(GO:0045494) |
| 0.0 |
1.0 |
GO:0040015 |
negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 |
1.8 |
GO:0000060 |
protein import into nucleus, translocation(GO:0000060) |
| 0.0 |
0.1 |
GO:1904749 |
protein localization to nucleolus(GO:1902570) regulation of protein localization to nucleolus(GO:1904749) positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.0 |
0.2 |
GO:0019695 |
choline metabolic process(GO:0019695) |
| 0.0 |
1.2 |
GO:0051310 |
ventricular system development(GO:0021591) metaphase plate congression(GO:0051310) |
| 0.0 |
1.2 |
GO:0061003 |
positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 |
0.1 |
GO:0015793 |
glycerol transport(GO:0015793) |
| 0.0 |
0.3 |
GO:1904262 |
negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 |
0.1 |
GO:0043652 |
interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) engulfment of apoptotic cell(GO:0043652) |
| 0.0 |
0.4 |
GO:0035493 |
SNARE complex assembly(GO:0035493) |
| 0.0 |
0.9 |
GO:0000266 |
mitochondrial fission(GO:0000266) |
| 0.0 |
0.1 |
GO:0051036 |
regulation of endosome size(GO:0051036) |
| 0.0 |
0.3 |
GO:0032516 |
positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 |
0.1 |
GO:1902268 |
negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 |
0.2 |
GO:0032926 |
negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 |
0.1 |
GO:0046602 |
regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 |
0.2 |
GO:0071318 |
cellular response to ATP(GO:0071318) |
| 0.0 |
0.8 |
GO:0031114 |
negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.0 |
0.5 |
GO:0006471 |
protein ADP-ribosylation(GO:0006471) |
| 0.0 |
0.2 |
GO:0042136 |
neurotransmitter biosynthetic process(GO:0042136) |
| 0.0 |
0.4 |
GO:0045103 |
intermediate filament-based process(GO:0045103) |
| 0.0 |
0.3 |
GO:0046855 |
phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 |
0.2 |
GO:0031017 |
exocrine pancreas development(GO:0031017) |
| 0.0 |
0.1 |
GO:1901165 |
positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 |
0.6 |
GO:0070232 |
regulation of T cell apoptotic process(GO:0070232) |
| 0.0 |
0.3 |
GO:0001574 |
ganglioside biosynthetic process(GO:0001574) |
| 0.0 |
0.0 |
GO:0006434 |
seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 |
0.4 |
GO:0001523 |
retinoid metabolic process(GO:0001523) |
| 0.0 |
0.0 |
GO:0002636 |
positive regulation of germinal center formation(GO:0002636) |
| 0.0 |
0.5 |
GO:0032611 |
interleukin-1 beta production(GO:0032611) |
| 0.0 |
0.2 |
GO:1990403 |
embryonic brain development(GO:1990403) |
| 0.0 |
0.2 |
GO:0051894 |
positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 |
0.1 |
GO:0071361 |
cellular response to ethanol(GO:0071361) |
| 0.0 |
0.1 |
GO:0060766 |
negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.0 |
0.2 |
GO:0009083 |
branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 |
0.2 |
GO:0016180 |
snRNA processing(GO:0016180) |
| 0.0 |
0.2 |
GO:0016486 |
peptide hormone processing(GO:0016486) |