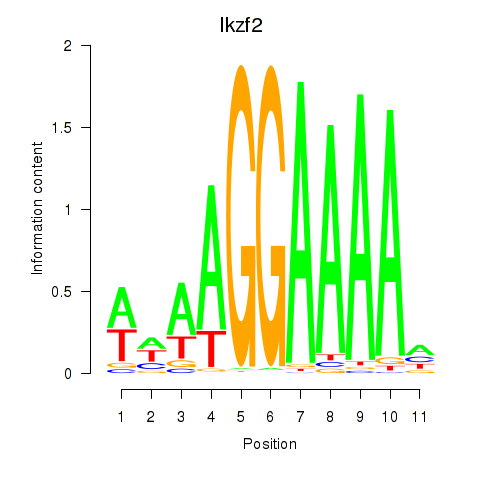
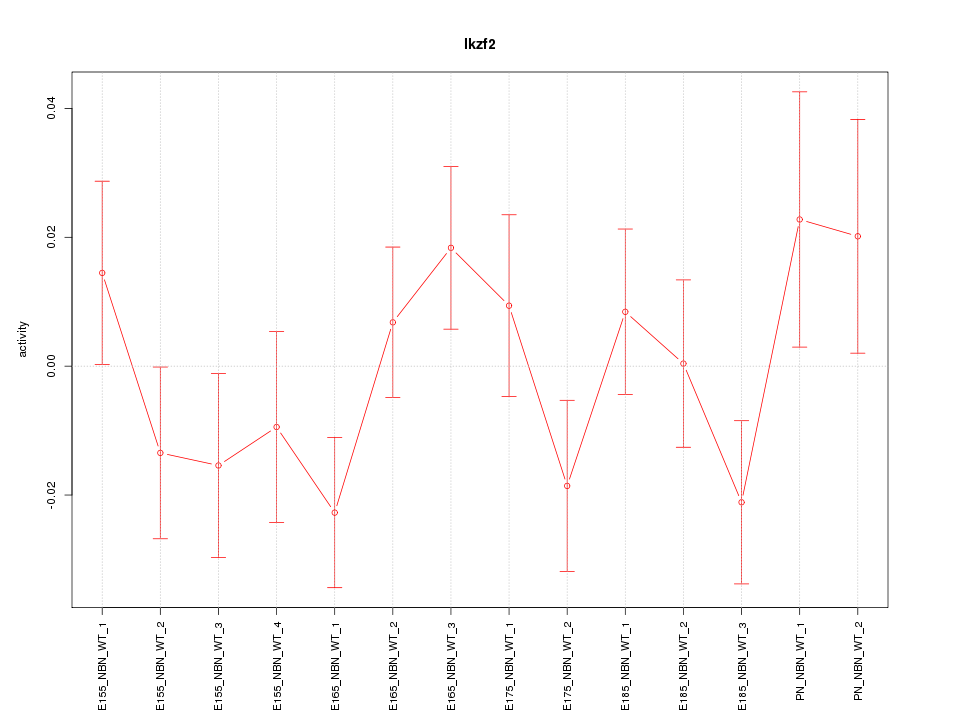
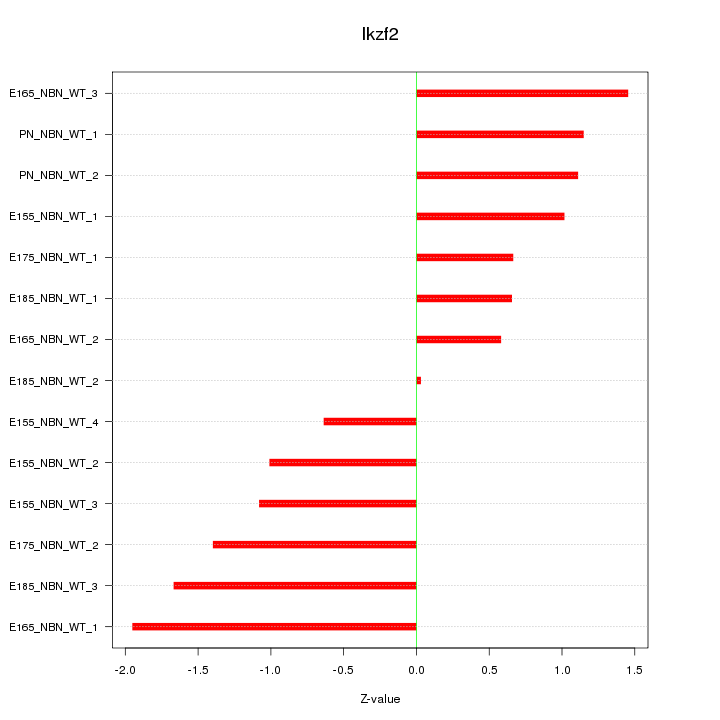
| 1.1 |
7.4 |
GO:0001887 |
selenium compound metabolic process(GO:0001887) |
| 0.6 |
2.9 |
GO:0060414 |
aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.6 |
1.7 |
GO:0021893 |
cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 0.4 |
1.2 |
GO:1903225 |
negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.3 |
0.9 |
GO:0090258 |
negative regulation of mitochondrial fission(GO:0090258) |
| 0.3 |
0.5 |
GO:0060838 |
lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.2 |
1.0 |
GO:1903026 |
negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.2 |
0.9 |
GO:0003199 |
endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) positive regulation of blood vessel remodeling(GO:2000504) |
| 0.2 |
0.9 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |
| 0.2 |
1.9 |
GO:0060501 |
positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.1 |
1.2 |
GO:0009404 |
toxin metabolic process(GO:0009404) |
| 0.1 |
1.3 |
GO:0061299 |
retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 |
1.1 |
GO:0061032 |
visceral serous pericardium development(GO:0061032) |
| 0.1 |
0.2 |
GO:0007161 |
calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.1 |
0.3 |
GO:0071726 |
response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) |
| 0.1 |
0.4 |
GO:0045578 |
negative regulation of B cell differentiation(GO:0045578) |
| 0.1 |
0.5 |
GO:0046477 |
glycosylceramide catabolic process(GO:0046477) |
| 0.1 |
0.8 |
GO:0097067 |
cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 |
0.1 |
GO:2000040 |
regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.1 |
0.4 |
GO:0035469 |
determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.1 |
0.6 |
GO:0031441 |
negative regulation of mRNA 3'-end processing(GO:0031441) |
| 0.1 |
0.3 |
GO:0040032 |
post-embryonic body morphogenesis(GO:0040032) |
| 0.1 |
0.2 |
GO:0046013 |
regulation of T cell homeostatic proliferation(GO:0046013) positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 |
0.3 |
GO:0021913 |
regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 |
0.5 |
GO:0042447 |
hormone catabolic process(GO:0042447) |
| 0.0 |
0.3 |
GO:0060312 |
regulation of blood vessel remodeling(GO:0060312) |
| 0.0 |
0.1 |
GO:0000821 |
regulation of arginine metabolic process(GO:0000821) |
| 0.0 |
1.0 |
GO:0045662 |
negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 |
1.4 |
GO:0001937 |
negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.0 |
0.2 |
GO:0034080 |
CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 |
0.2 |
GO:0006287 |
base-excision repair, gap-filling(GO:0006287) |
| 0.0 |
1.2 |
GO:0051642 |
centrosome localization(GO:0051642) |
| 0.0 |
0.4 |
GO:0031017 |
exocrine pancreas development(GO:0031017) |
| 0.0 |
0.2 |
GO:0048625 |
myoblast fate commitment(GO:0048625) |
| 0.0 |
0.2 |
GO:0007258 |
JUN phosphorylation(GO:0007258) |
| 0.0 |
0.6 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.0 |
0.5 |
GO:0060008 |
Sertoli cell differentiation(GO:0060008) |
| 0.0 |
0.2 |
GO:0006883 |
cellular sodium ion homeostasis(GO:0006883) |
| 0.0 |
0.0 |
GO:0072050 |
comma-shaped body morphogenesis(GO:0072049) S-shaped body morphogenesis(GO:0072050) |
| 0.0 |
0.2 |
GO:0051574 |
positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 |
0.4 |
GO:0010842 |
retina layer formation(GO:0010842) |
| 0.0 |
0.1 |
GO:0098532 |
histone H3-K27 trimethylation(GO:0098532) |
| 0.0 |
0.1 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 |
2.1 |
GO:0043062 |
extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |