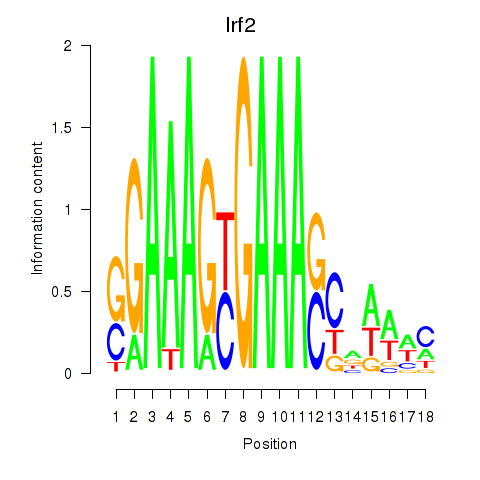
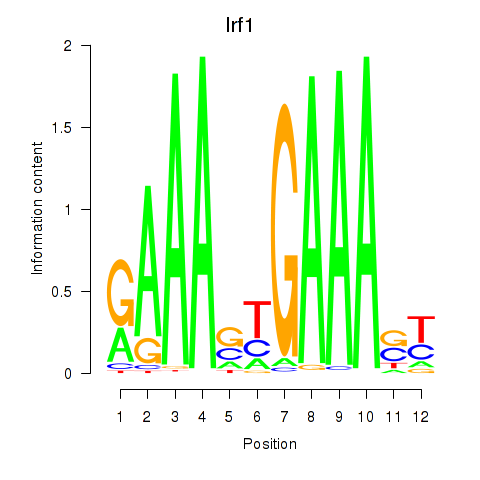
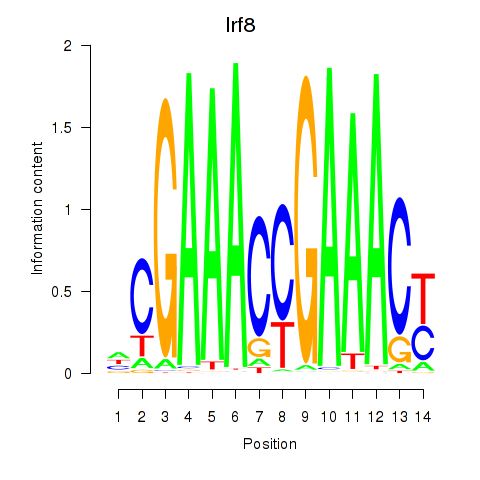
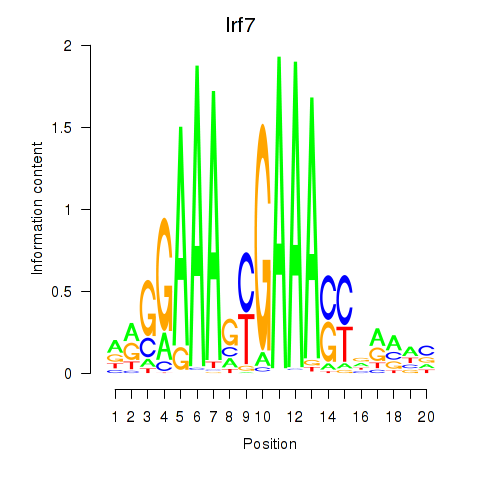
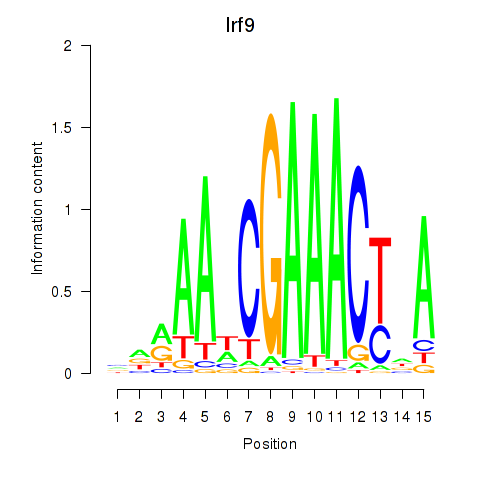
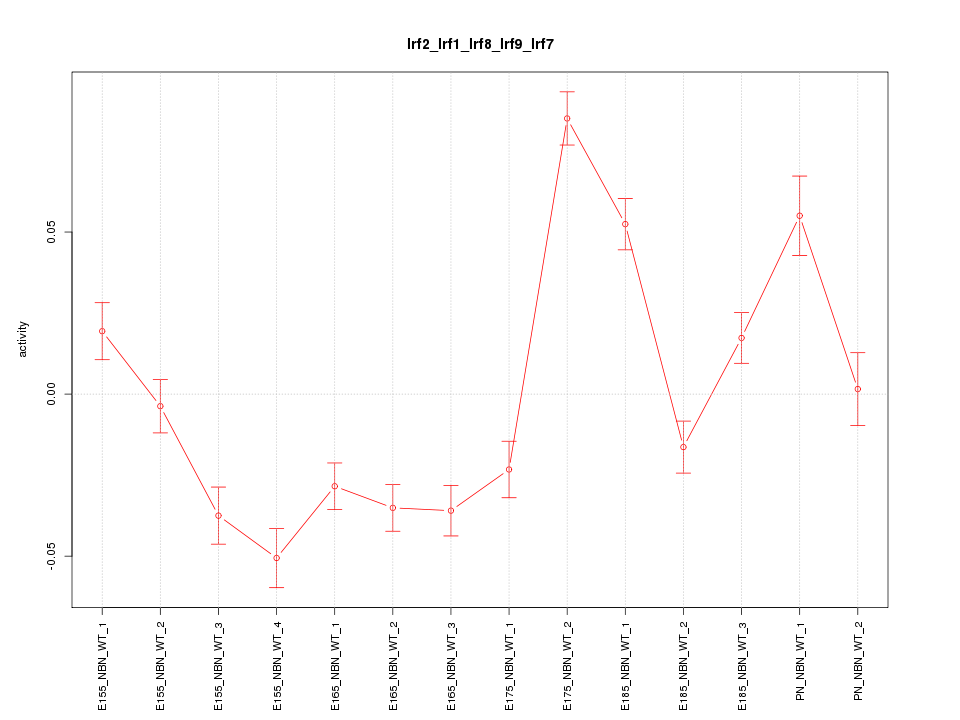
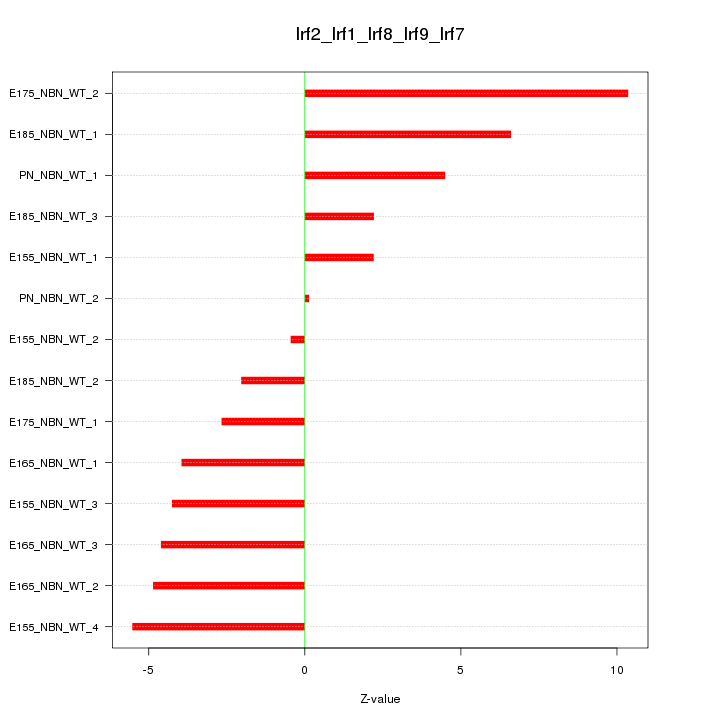
| 3.8 |
11.4 |
GO:2000562 |
negative regulation of interferon-gamma secretion(GO:1902714) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 3.7 |
14.6 |
GO:1901252 |
regulation of intracellular transport of viral material(GO:1901252) |
| 3.5 |
14.2 |
GO:0009597 |
detection of virus(GO:0009597) |
| 3.4 |
10.3 |
GO:0032687 |
negative regulation of interferon-alpha production(GO:0032687) |
| 3.1 |
9.2 |
GO:0001580 |
detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 3.0 |
14.8 |
GO:1900122 |
positive regulation of receptor binding(GO:1900122) |
| 1.6 |
4.7 |
GO:1902915 |
negative regulation of protein import into nucleus, translocation(GO:0033159) protein poly-ADP-ribosylation(GO:0070212) negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 1.1 |
4.4 |
GO:2001181 |
positive regulation of interleukin-10 secretion(GO:2001181) |
| 1.0 |
7.9 |
GO:0032020 |
ISG15-protein conjugation(GO:0032020) |
| 0.9 |
3.7 |
GO:0002606 |
positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.8 |
2.5 |
GO:0070944 |
neutrophil mediated killing of bacterium(GO:0070944) |
| 0.7 |
2.2 |
GO:0010533 |
regulation of activation of Janus kinase activity(GO:0010533) regulation of activation of JAK2 kinase activity(GO:0010534) |
| 0.7 |
10.8 |
GO:2001275 |
positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.7 |
2.2 |
GO:0033566 |
gamma-tubulin complex localization(GO:0033566) |
| 0.7 |
12.6 |
GO:0042832 |
defense response to protozoan(GO:0042832) |
| 0.7 |
2.7 |
GO:1903984 |
positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.7 |
2.0 |
GO:0042196 |
dichloromethane metabolic process(GO:0018900) chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 0.6 |
1.8 |
GO:0070269 |
pyroptosis(GO:0070269) |
| 0.6 |
2.4 |
GO:0035934 |
corticosterone secretion(GO:0035934) |
| 0.5 |
2.2 |
GO:0003430 |
tolerance induction to self antigen(GO:0002513) growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.5 |
1.6 |
GO:0072092 |
ureteric bud invasion(GO:0072092) |
| 0.5 |
3.1 |
GO:0003383 |
apical constriction(GO:0003383) |
| 0.5 |
1.6 |
GO:0007210 |
serotonin receptor signaling pathway(GO:0007210) |
| 0.5 |
1.6 |
GO:0039530 |
MDA-5 signaling pathway(GO:0039530) |
| 0.5 |
2.6 |
GO:0070460 |
thyroid-stimulating hormone secretion(GO:0070460) |
| 0.5 |
1.0 |
GO:0050713 |
negative regulation of interleukin-1 secretion(GO:0050711) negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.5 |
7.5 |
GO:0010447 |
response to acidic pH(GO:0010447) |
| 0.5 |
1.9 |
GO:0044565 |
dendritic cell proliferation(GO:0044565) |
| 0.5 |
3.3 |
GO:0042590 |
antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.5 |
1.4 |
GO:0046098 |
guanine metabolic process(GO:0046098) |
| 0.5 |
1.4 |
GO:1904139 |
microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.4 |
1.3 |
GO:1990481 |
snoRNA guided rRNA pseudouridine synthesis(GO:0000454) snRNA pseudouridine synthesis(GO:0031120) mRNA pseudouridine synthesis(GO:1990481) |
| 0.4 |
2.6 |
GO:1902036 |
regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.4 |
2.2 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.4 |
0.9 |
GO:0032701 |
negative regulation of interleukin-18 production(GO:0032701) |
| 0.4 |
10.6 |
GO:0035456 |
response to interferon-beta(GO:0035456) |
| 0.4 |
1.8 |
GO:1900170 |
negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.4 |
1.1 |
GO:0061537 |
glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.4 |
0.7 |
GO:0038091 |
VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) |
| 0.4 |
2.8 |
GO:0060665 |
regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.4 |
0.4 |
GO:0032621 |
interleukin-18 production(GO:0032621) |
| 0.4 |
2.5 |
GO:0031666 |
positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.3 |
2.8 |
GO:0048625 |
myoblast fate commitment(GO:0048625) |
| 0.3 |
1.0 |
GO:0090272 |
negative regulation of fibroblast growth factor production(GO:0090272) |
| 0.3 |
2.4 |
GO:0090336 |
positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.3 |
0.7 |
GO:1903225 |
negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.3 |
1.3 |
GO:2000313 |
fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.3 |
1.0 |
GO:0001951 |
intestinal D-glucose absorption(GO:0001951) terminal web assembly(GO:1902896) |
| 0.3 |
4.6 |
GO:0050930 |
induction of positive chemotaxis(GO:0050930) |
| 0.3 |
9.7 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.3 |
0.9 |
GO:0006059 |
hexitol metabolic process(GO:0006059) alditol biosynthetic process(GO:0019401) |
| 0.3 |
0.9 |
GO:0015889 |
cobalamin transport(GO:0015889) |
| 0.3 |
0.9 |
GO:2000668 |
dendritic cell apoptotic process(GO:0097048) regulation of dendritic cell apoptotic process(GO:2000668) |
| 0.3 |
0.9 |
GO:0048162 |
preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.3 |
1.7 |
GO:0032488 |
Cdc42 protein signal transduction(GO:0032488) |
| 0.3 |
0.8 |
GO:0001866 |
NK T cell proliferation(GO:0001866) |
| 0.3 |
1.6 |
GO:0007258 |
JUN phosphorylation(GO:0007258) |
| 0.3 |
1.6 |
GO:0035989 |
tendon development(GO:0035989) |
| 0.3 |
0.8 |
GO:0033860 |
regulation of NAD(P)H oxidase activity(GO:0033860) |
| 0.3 |
1.3 |
GO:0034421 |
post-translational protein acetylation(GO:0034421) |
| 0.2 |
0.5 |
GO:0008065 |
establishment of blood-nerve barrier(GO:0008065) |
| 0.2 |
1.8 |
GO:0071360 |
cellular response to exogenous dsRNA(GO:0071360) |
| 0.2 |
1.1 |
GO:0010032 |
meiotic chromosome condensation(GO:0010032) |
| 0.2 |
0.2 |
GO:2000564 |
CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.2 |
1.1 |
GO:0045918 |
negative regulation of cytolysis(GO:0045918) |
| 0.2 |
1.4 |
GO:0019532 |
oxalate transport(GO:0019532) |
| 0.2 |
0.6 |
GO:0014891 |
striated muscle atrophy(GO:0014891) |
| 0.2 |
0.8 |
GO:0036462 |
TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.2 |
0.2 |
GO:1903463 |
regulation of mitotic cell cycle DNA replication(GO:1903463) |
| 0.2 |
0.6 |
GO:0048686 |
regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.2 |
0.4 |
GO:0003406 |
retinal pigment epithelium development(GO:0003406) |
| 0.2 |
0.9 |
GO:0010701 |
positive regulation of norepinephrine secretion(GO:0010701) |
| 0.2 |
0.7 |
GO:0048143 |
astrocyte activation(GO:0048143) |
| 0.2 |
0.7 |
GO:0032901 |
positive regulation of neurotrophin production(GO:0032901) |
| 0.2 |
5.0 |
GO:0045071 |
negative regulation of viral genome replication(GO:0045071) |
| 0.2 |
0.7 |
GO:1902202 |
regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.2 |
0.7 |
GO:0051151 |
negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.2 |
0.5 |
GO:1904457 |
positive regulation of neuronal action potential(GO:1904457) |
| 0.2 |
2.5 |
GO:0042759 |
long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.2 |
0.5 |
GO:0043456 |
regulation of pentose-phosphate shunt(GO:0043456) |
| 0.2 |
1.2 |
GO:0060842 |
arterial endothelial cell differentiation(GO:0060842) |
| 0.2 |
0.6 |
GO:0006447 |
regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 0.2 |
0.5 |
GO:0051342 |
regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.2 |
0.6 |
GO:1902626 |
assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 |
1.6 |
GO:0042572 |
retinol metabolic process(GO:0042572) |
| 0.1 |
5.0 |
GO:0071346 |
cellular response to interferon-gamma(GO:0071346) |
| 0.1 |
2.1 |
GO:0034501 |
protein localization to kinetochore(GO:0034501) |
| 0.1 |
3.3 |
GO:0016082 |
synaptic vesicle priming(GO:0016082) |
| 0.1 |
0.7 |
GO:0046598 |
positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 |
6.0 |
GO:0046825 |
regulation of protein export from nucleus(GO:0046825) |
| 0.1 |
1.7 |
GO:0044458 |
motile cilium assembly(GO:0044458) |
| 0.1 |
0.4 |
GO:0006553 |
lysine metabolic process(GO:0006553) |
| 0.1 |
1.1 |
GO:0031953 |
negative regulation of protein autophosphorylation(GO:0031953) |
| 0.1 |
0.8 |
GO:0032483 |
regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 |
1.5 |
GO:0097067 |
cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 |
1.5 |
GO:0021527 |
spinal cord association neuron differentiation(GO:0021527) |
| 0.1 |
0.5 |
GO:0031509 |
telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.1 |
0.8 |
GO:0042795 |
snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 |
0.9 |
GO:0045779 |
negative regulation of bone resorption(GO:0045779) |
| 0.1 |
0.5 |
GO:0032511 |
late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 |
2.1 |
GO:0045292 |
mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 |
2.2 |
GO:0051044 |
positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 |
2.6 |
GO:0035855 |
megakaryocyte development(GO:0035855) |
| 0.1 |
0.4 |
GO:2000338 |
positive regulation of interleukin-23 production(GO:0032747) chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.1 |
0.7 |
GO:1900029 |
positive regulation of ruffle assembly(GO:1900029) |
| 0.1 |
1.2 |
GO:0032876 |
negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 |
0.6 |
GO:0072383 |
plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 |
0.9 |
GO:0019348 |
dolichol metabolic process(GO:0019348) |
| 0.1 |
1.0 |
GO:1902902 |
negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 |
0.5 |
GO:0042758 |
long-chain fatty acid catabolic process(GO:0042758) |
| 0.1 |
0.5 |
GO:0097116 |
gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.1 |
0.6 |
GO:0060762 |
regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.1 |
1.2 |
GO:0046321 |
positive regulation of fatty acid oxidation(GO:0046321) |
| 0.1 |
1.1 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 |
0.4 |
GO:0051132 |
NK T cell activation(GO:0051132) |
| 0.1 |
0.6 |
GO:0006108 |
malate metabolic process(GO:0006108) |
| 0.1 |
1.2 |
GO:0070914 |
UV-damage excision repair(GO:0070914) |
| 0.1 |
0.7 |
GO:0097039 |
protein linear polyubiquitination(GO:0097039) |
| 0.1 |
10.8 |
GO:0007286 |
spermatid development(GO:0007286) |
| 0.1 |
1.0 |
GO:1903608 |
protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 |
1.6 |
GO:0050829 |
defense response to Gram-negative bacterium(GO:0050829) |
| 0.1 |
0.2 |
GO:0006990 |
positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.1 |
1.9 |
GO:1901522 |
positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.1 |
0.4 |
GO:1903887 |
motile primary cilium assembly(GO:1903887) |
| 0.1 |
0.3 |
GO:0002314 |
germinal center B cell differentiation(GO:0002314) |
| 0.1 |
0.3 |
GO:1902990 |
mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.1 |
0.3 |
GO:1904569 |
regulation of selenocysteine incorporation(GO:1904569) |
| 0.1 |
0.6 |
GO:0015840 |
urea transport(GO:0015840) urea transmembrane transport(GO:0071918) |
| 0.1 |
1.1 |
GO:0034497 |
protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 |
0.3 |
GO:0072602 |
interleukin-4 secretion(GO:0072602) |
| 0.1 |
0.6 |
GO:2000194 |
regulation of female gonad development(GO:2000194) |
| 0.1 |
1.4 |
GO:2000310 |
regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.1 |
0.5 |
GO:1901162 |
serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.1 |
0.4 |
GO:0046501 |
protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 |
1.4 |
GO:0043552 |
positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.1 |
0.3 |
GO:0036481 |
intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036481) regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.1 |
0.4 |
GO:0090050 |
positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 |
0.3 |
GO:0005981 |
regulation of glycogen catabolic process(GO:0005981) |
| 0.1 |
0.6 |
GO:2000781 |
positive regulation of double-strand break repair(GO:2000781) |
| 0.1 |
0.1 |
GO:0060340 |
positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.1 |
7.3 |
GO:0016052 |
carbohydrate catabolic process(GO:0016052) |
| 0.1 |
1.8 |
GO:0043171 |
peptide catabolic process(GO:0043171) |
| 0.1 |
0.4 |
GO:0002347 |
response to tumor cell(GO:0002347) |
| 0.1 |
0.9 |
GO:0045662 |
negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 |
0.8 |
GO:0000188 |
inactivation of MAPK activity(GO:0000188) |
| 0.1 |
0.7 |
GO:1904262 |
negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 |
0.1 |
GO:0006624 |
vacuolar protein processing(GO:0006624) |
| 0.1 |
2.5 |
GO:0042632 |
cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.1 |
0.3 |
GO:0060040 |
retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 |
0.3 |
GO:1902261 |
positive regulation of delayed rectifier potassium channel activity(GO:1902261) positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.1 |
0.2 |
GO:0060923 |
cardiac muscle cell fate commitment(GO:0060923) |
| 0.1 |
0.2 |
GO:0000821 |
regulation of arginine metabolic process(GO:0000821) |
| 0.1 |
0.2 |
GO:1904879 |
positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.1 |
0.1 |
GO:0018307 |
enzyme active site formation(GO:0018307) |
| 0.1 |
0.5 |
GO:1904587 |
glycoprotein ERAD pathway(GO:0097466) response to glycoprotein(GO:1904587) |
| 0.1 |
1.0 |
GO:0031115 |
negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 |
0.2 |
GO:0038018 |
Wnt receptor catabolic process(GO:0038018) |
| 0.0 |
0.8 |
GO:0006744 |
ubiquinone biosynthetic process(GO:0006744) |
| 0.0 |
0.9 |
GO:0007194 |
negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 |
0.8 |
GO:0001963 |
synaptic transmission, dopaminergic(GO:0001963) |
| 0.0 |
0.1 |
GO:0044778 |
meiotic DNA integrity checkpoint(GO:0044778) |
| 0.0 |
0.5 |
GO:0072010 |
renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 |
0.3 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 |
1.4 |
GO:0034724 |
DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 |
0.2 |
GO:0071028 |
nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 |
0.1 |
GO:2000301 |
negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 |
0.4 |
GO:0030539 |
male genitalia development(GO:0030539) |
| 0.0 |
0.2 |
GO:0032815 |
negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 |
0.6 |
GO:0000183 |
chromatin silencing at rDNA(GO:0000183) |
| 0.0 |
1.0 |
GO:0007638 |
mechanosensory behavior(GO:0007638) |
| 0.0 |
0.3 |
GO:0060613 |
fat pad development(GO:0060613) |
| 0.0 |
0.2 |
GO:0046784 |
viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 |
0.1 |
GO:0019085 |
early viral transcription(GO:0019085) late viral transcription(GO:0019086) |
| 0.0 |
1.3 |
GO:0090162 |
establishment of epithelial cell polarity(GO:0090162) |
| 0.0 |
0.3 |
GO:0032534 |
regulation of microvillus assembly(GO:0032534) |
| 0.0 |
0.2 |
GO:0006041 |
glucosamine metabolic process(GO:0006041) |
| 0.0 |
0.2 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 |
0.3 |
GO:0042996 |
regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 |
1.7 |
GO:0035019 |
somatic stem cell population maintenance(GO:0035019) |
| 0.0 |
0.2 |
GO:1903025 |
regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 |
0.1 |
GO:0036297 |
interstrand cross-link repair(GO:0036297) |
| 0.0 |
0.4 |
GO:0019985 |
translesion synthesis(GO:0019985) |
| 0.0 |
0.2 |
GO:0017196 |
N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 |
0.1 |
GO:0071492 |
cellular response to UV-A(GO:0071492) |
| 0.0 |
0.9 |
GO:0046677 |
response to antibiotic(GO:0046677) |
| 0.0 |
0.2 |
GO:0051096 |
regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.0 |
0.3 |
GO:1901970 |
positive regulation of mitotic sister chromatid separation(GO:1901970) |
| 0.0 |
0.3 |
GO:0044539 |
long-chain fatty acid import(GO:0044539) |
| 0.0 |
0.2 |
GO:0010587 |
miRNA catabolic process(GO:0010587) |
| 0.0 |
0.2 |
GO:0032836 |
glomerular basement membrane development(GO:0032836) |
| 0.0 |
0.1 |
GO:2000383 |
regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.0 |
0.5 |
GO:2000001 |
regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 |
0.3 |
GO:1904293 |
negative regulation of ERAD pathway(GO:1904293) |
| 0.0 |
0.8 |
GO:0051642 |
centrosome localization(GO:0051642) |
| 0.0 |
0.6 |
GO:0016180 |
snRNA processing(GO:0016180) |
| 0.0 |
0.2 |
GO:1902306 |
negative regulation of sodium ion transmembrane transport(GO:1902306) |
| 0.0 |
0.1 |
GO:0014012 |
peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 |
0.2 |
GO:0000291 |
nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.0 |
0.2 |
GO:0051984 |
positive regulation of chromosome segregation(GO:0051984) |
| 0.0 |
0.1 |
GO:0090310 |
negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.0 |
0.3 |
GO:0050434 |
positive regulation of viral transcription(GO:0050434) |
| 0.0 |
0.1 |
GO:0034975 |
protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 |
0.1 |
GO:0044375 |
regulation of peroxisome size(GO:0044375) |
| 0.0 |
0.2 |
GO:0000491 |
small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 |
4.1 |
GO:0043062 |
extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 |
0.8 |
GO:0007032 |
endosome organization(GO:0007032) |
| 0.0 |
1.3 |
GO:0006749 |
glutathione metabolic process(GO:0006749) |
| 0.0 |
1.1 |
GO:0006289 |
nucleotide-excision repair(GO:0006289) |
| 0.0 |
2.4 |
GO:0016579 |
protein deubiquitination(GO:0016579) |
| 0.0 |
0.2 |
GO:0009143 |
nucleoside triphosphate catabolic process(GO:0009143) |
| 0.0 |
0.8 |
GO:0005978 |
glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 |
0.1 |
GO:0002710 |
negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 |
0.3 |
GO:0006958 |
complement activation, classical pathway(GO:0006958) |
| 0.0 |
0.4 |
GO:0031365 |
N-terminal protein amino acid modification(GO:0031365) |
| 0.0 |
0.1 |
GO:2000348 |
regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 |
0.1 |
GO:1904424 |
regulation of GTP binding(GO:1904424) |
| 0.0 |
0.0 |
GO:0001771 |
immunological synapse formation(GO:0001771) |
| 0.0 |
0.2 |
GO:0006388 |
tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 |
4.5 |
GO:0006914 |
autophagy(GO:0006914) |
| 0.0 |
0.2 |
GO:0045899 |
positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 |
0.1 |
GO:0007064 |
mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 |
0.2 |
GO:0016926 |
protein desumoylation(GO:0016926) |
| 0.0 |
0.1 |
GO:2000172 |
regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.0 |
1.1 |
GO:0007605 |
sensory perception of sound(GO:0007605) |
| 0.0 |
0.2 |
GO:0043516 |
regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043516) |
| 0.0 |
0.1 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 |
0.1 |
GO:0006122 |
mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 |
0.8 |
GO:0050885 |
neuromuscular process controlling balance(GO:0050885) |
| 0.0 |
0.0 |
GO:0048318 |
axial mesoderm development(GO:0048318) |
| 0.0 |
0.1 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 |
0.1 |
GO:0031584 |
activation of phospholipase D activity(GO:0031584) |
| 0.0 |
0.9 |
GO:0007050 |
cell cycle arrest(GO:0007050) |
| 0.0 |
0.1 |
GO:0008340 |
determination of adult lifespan(GO:0008340) |
| 0.0 |
0.0 |
GO:0002827 |
regulation of T-helper 1 type immune response(GO:0002825) positive regulation of T-helper 1 type immune response(GO:0002827) regulation of T-helper 1 cell differentiation(GO:0045625) positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.0 |
0.0 |
GO:0097211 |
prolactin secretion(GO:0070459) response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |