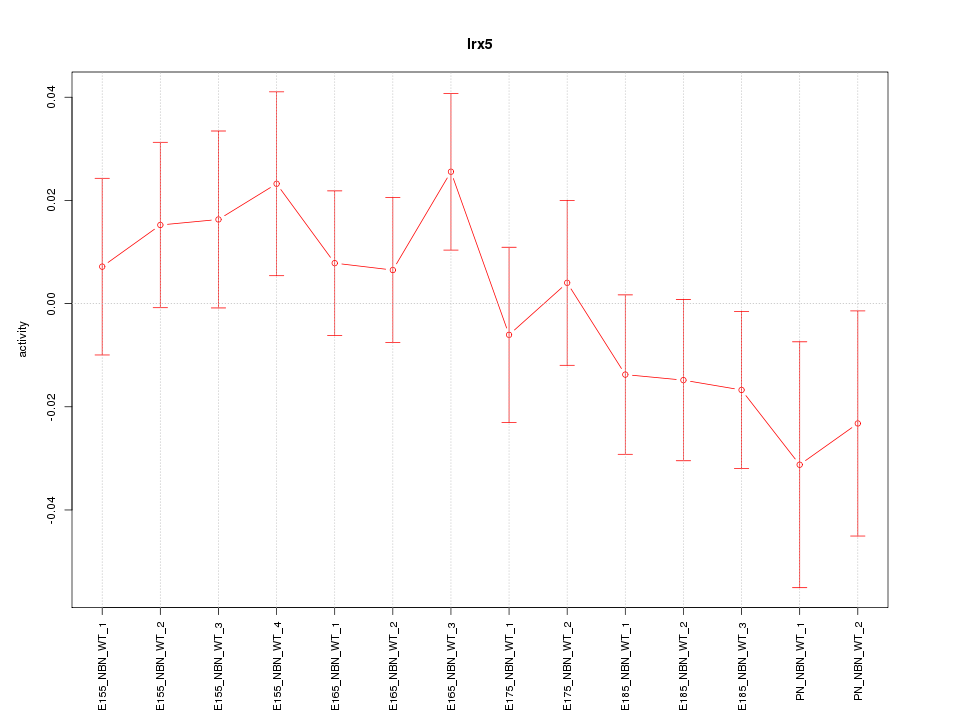
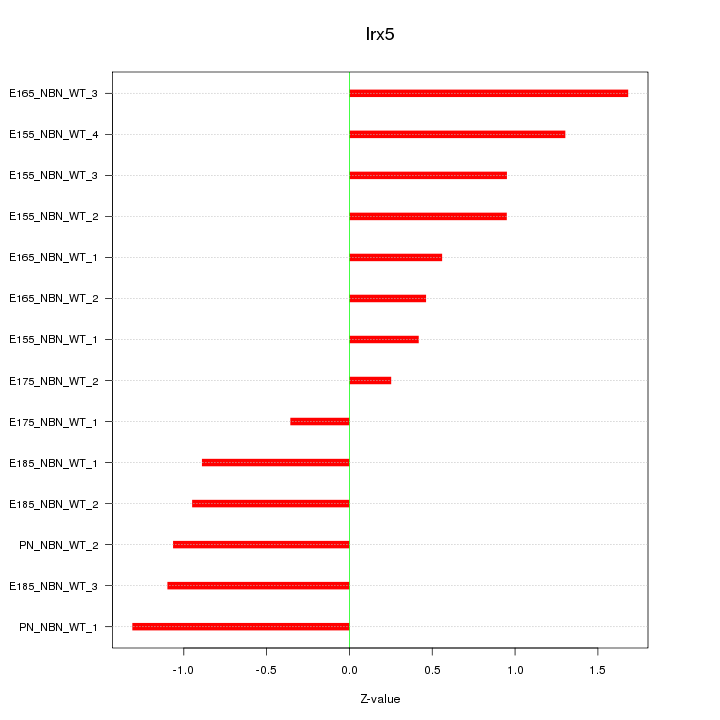
Motif ID: Irx5
Z-value: 0.963
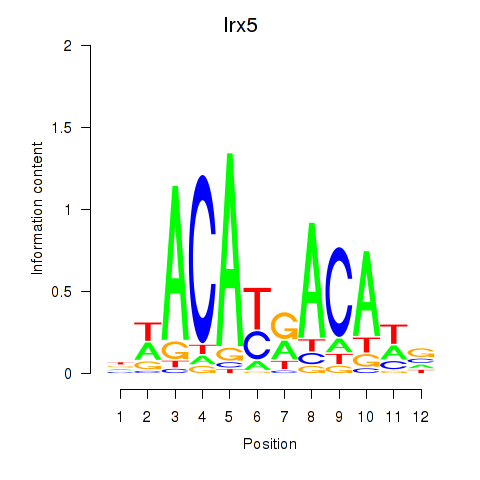
Transcription factors associated with Irx5:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Irx5 | ENSMUSG00000031737.10 | Irx5 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Irx5 | mm10_v2_chr8_+_92357787_92357801 | 0.56 | 3.8e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:1904058 | positive regulation of sensory perception of pain(GO:1904058) |
| 0.4 | 3.3 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.3 | 1.0 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.2 | 1.6 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.2 | 0.8 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.2 | 2.1 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.2 | 3.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.2 | 0.5 | GO:1902524 | positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.2 | 0.5 | GO:0072425 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.1 | 1.3 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 1.2 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.5 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 0.4 | GO:0010986 | complement receptor mediated signaling pathway(GO:0002430) GPI anchor release(GO:0006507) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.1 | 0.9 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.1 | 0.3 | GO:1905154 | negative regulation of tumor necrosis factor secretion(GO:1904468) negative regulation of membrane invagination(GO:1905154) |
| 0.1 | 0.4 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 | 0.9 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 1.1 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.8 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.1 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.0 | 0.7 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.5 | GO:0007566 | embryo implantation(GO:0007566) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.2 | 3.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.2 | 1.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 0.5 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.1 | 0.5 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.1 | 1.3 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 0.4 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.7 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 0.8 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 1.2 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 2.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.3 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.5 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 1.1 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 2.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 2.1 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.8 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.2 | GO:0005865 | striated muscle thin filament(GO:0005865) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.3 | 1.0 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.3 | 1.6 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.2 | 1.3 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 2.0 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.5 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 2.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 1.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 3.3 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.4 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 1.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.5 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 2.1 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |