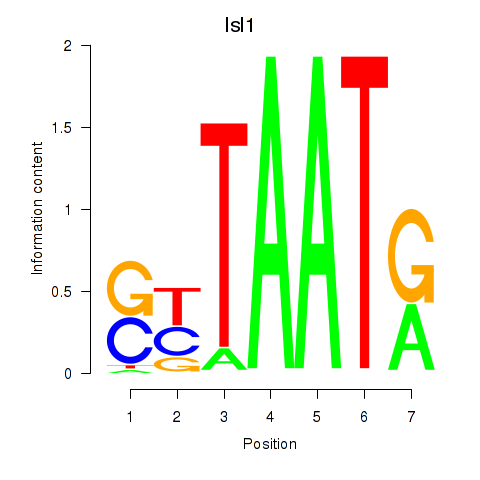
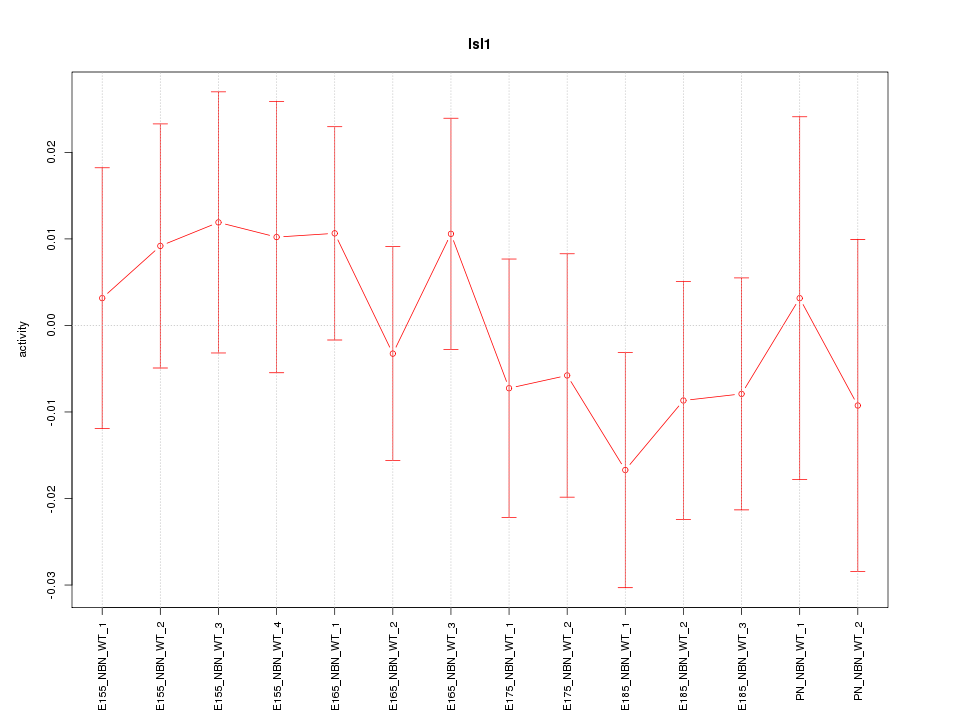
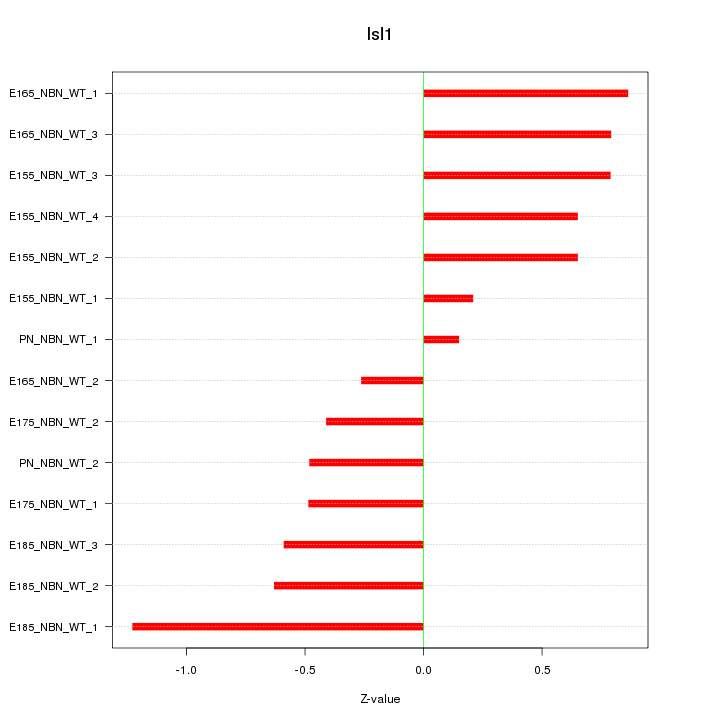
| 0.9 |
2.7 |
GO:0051892 |
negative regulation of cardioblast differentiation(GO:0051892) |
| 0.8 |
6.6 |
GO:0060164 |
regulation of timing of neuron differentiation(GO:0060164) |
| 0.4 |
2.4 |
GO:1904936 |
forebrain anterior/posterior pattern specification(GO:0021797) cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.2 |
0.5 |
GO:0060023 |
noradrenergic neuron differentiation(GO:0003357) soft palate development(GO:0060023) |
| 0.2 |
0.5 |
GO:0042320 |
regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) circadian sleep/wake cycle, REM sleep(GO:0042747) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.1 |
0.4 |
GO:1901731 |
calcium-mediated signaling using extracellular calcium source(GO:0035585) positive regulation of platelet aggregation(GO:1901731) |
| 0.1 |
0.6 |
GO:0048245 |
eosinophil chemotaxis(GO:0048245) |
| 0.1 |
1.2 |
GO:0046543 |
development of secondary female sexual characteristics(GO:0046543) |
| 0.1 |
0.5 |
GO:2000253 |
positive regulation of feeding behavior(GO:2000253) |
| 0.1 |
0.4 |
GO:0014846 |
esophagus smooth muscle contraction(GO:0014846) |
| 0.1 |
1.0 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.1 |
1.6 |
GO:2001224 |
positive regulation of neuron migration(GO:2001224) |
| 0.1 |
1.0 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 |
1.0 |
GO:0021819 |
layer formation in cerebral cortex(GO:0021819) |
| 0.1 |
0.4 |
GO:0001880 |
Mullerian duct regression(GO:0001880) osteoblast fate commitment(GO:0002051) |
| 0.1 |
0.6 |
GO:0090073 |
positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 |
0.3 |
GO:1903690 |
negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 |
0.5 |
GO:0036371 |
protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.1 |
0.2 |
GO:2000686 |
regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.1 |
1.3 |
GO:0006359 |
regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.1 |
0.2 |
GO:0060060 |
post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.0 |
0.1 |
GO:0072425 |
signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.0 |
0.1 |
GO:0021691 |
cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 |
0.3 |
GO:0014816 |
skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 |
0.2 |
GO:0043490 |
malate-aspartate shuttle(GO:0043490) |
| 0.0 |
0.1 |
GO:0046077 |
dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) UDP metabolic process(GO:0046048) dUDP metabolic process(GO:0046077) |
| 0.0 |
0.3 |
GO:0018230 |
peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 |
0.4 |
GO:0010650 |
positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 |
0.1 |
GO:0048312 |
intracellular distribution of mitochondria(GO:0048312) |
| 0.0 |
0.3 |
GO:2001256 |
regulation of store-operated calcium entry(GO:2001256) |
| 0.0 |
0.2 |
GO:0006570 |
tyrosine metabolic process(GO:0006570) |
| 0.0 |
0.2 |
GO:0071044 |
histone mRNA catabolic process(GO:0071044) |
| 0.0 |
0.1 |
GO:0001778 |
plasma membrane repair(GO:0001778) |
| 0.0 |
0.4 |
GO:0098743 |
cartilage condensation(GO:0001502) cell aggregation(GO:0098743) |
| 0.0 |
0.5 |
GO:0003301 |
physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 |
0.3 |
GO:1902043 |
positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 |
1.7 |
GO:0051028 |
mRNA transport(GO:0051028) |
| 0.0 |
0.2 |
GO:0010603 |
regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |