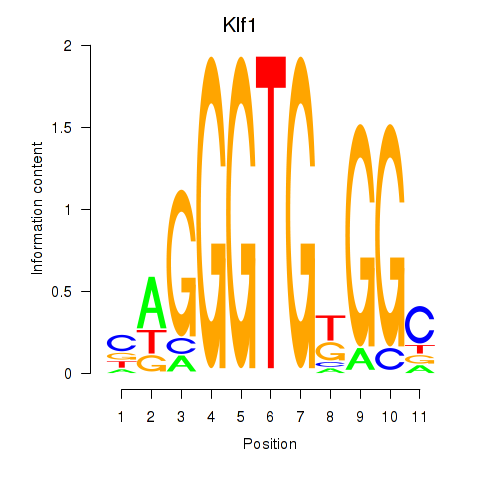
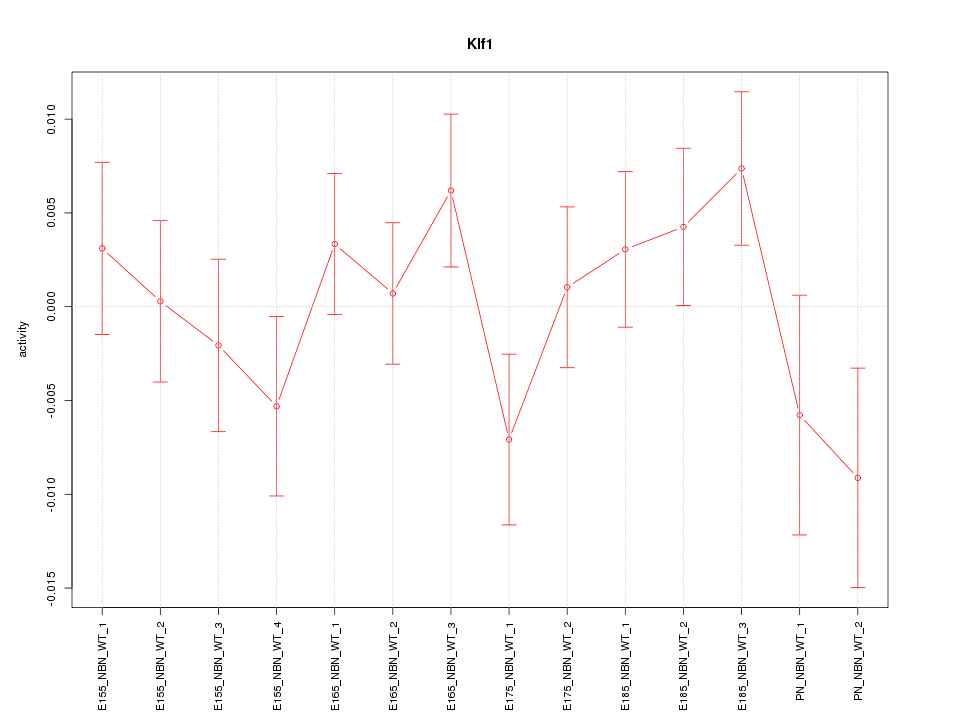
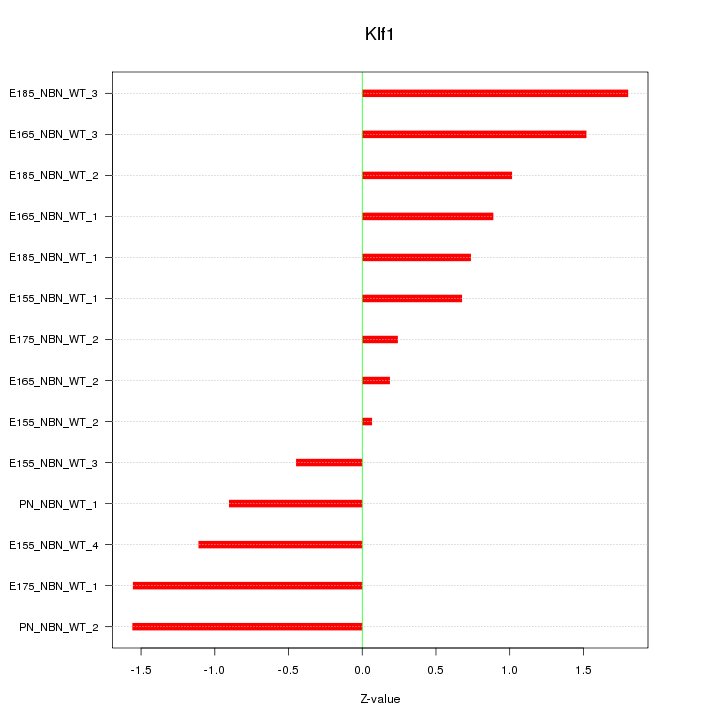
| 0.2 |
1.0 |
GO:1903553 |
positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.2 |
0.8 |
GO:0000379 |
tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.2 |
1.2 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.2 |
0.6 |
GO:0051902 |
negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.2 |
0.6 |
GO:0010986 |
positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.2 |
1.1 |
GO:0032796 |
uropod organization(GO:0032796) |
| 0.2 |
0.5 |
GO:0015772 |
disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.2 |
0.5 |
GO:0006682 |
galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.2 |
0.5 |
GO:1901731 |
calcium-mediated signaling using extracellular calcium source(GO:0035585) positive regulation of platelet aggregation(GO:1901731) |
| 0.2 |
0.6 |
GO:0007352 |
zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.2 |
0.8 |
GO:0019659 |
glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.2 |
0.6 |
GO:0042321 |
negative regulation of circadian sleep/wake cycle, sleep(GO:0042321) |
| 0.2 |
0.5 |
GO:0070172 |
positive regulation of tooth mineralization(GO:0070172) |
| 0.1 |
0.6 |
GO:0035606 |
peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 |
0.4 |
GO:0007621 |
negative regulation of female receptivity(GO:0007621) |
| 0.1 |
0.9 |
GO:0032804 |
negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) |
| 0.1 |
0.7 |
GO:1904721 |
regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.1 |
0.6 |
GO:0032275 |
luteinizing hormone secretion(GO:0032275) positive regulation of gonadotropin secretion(GO:0032278) |
| 0.1 |
0.3 |
GO:0021506 |
anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.1 |
0.4 |
GO:1903334 |
positive regulation of protein folding(GO:1903334) |
| 0.1 |
0.5 |
GO:0051708 |
intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) cytosol to ER transport(GO:0046967) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.1 |
0.4 |
GO:0090298 |
positive regulation of fat cell proliferation(GO:0070346) negative regulation of mitochondrial DNA replication(GO:0090298) |
| 0.1 |
0.9 |
GO:1904417 |
regulation of xenophagy(GO:1904415) positive regulation of xenophagy(GO:1904417) |
| 0.1 |
0.4 |
GO:0042536 |
negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.1 |
0.7 |
GO:0060017 |
parathyroid gland development(GO:0060017) |
| 0.1 |
0.5 |
GO:0060830 |
ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.1 |
0.4 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 |
0.3 |
GO:0051295 |
establishment of meiotic spindle localization(GO:0051295) formin-nucleated actin cable assembly(GO:0070649) |
| 0.1 |
0.3 |
GO:0046959 |
habituation(GO:0046959) |
| 0.1 |
0.6 |
GO:0048133 |
germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 |
0.1 |
GO:0060994 |
regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) |
| 0.1 |
1.0 |
GO:0033280 |
response to vitamin D(GO:0033280) |
| 0.1 |
0.3 |
GO:0030070 |
insulin processing(GO:0030070) |
| 0.1 |
0.3 |
GO:1903903 |
regulation of establishment of T cell polarity(GO:1903903) |
| 0.1 |
1.1 |
GO:0090084 |
negative regulation of inclusion body assembly(GO:0090084) |
| 0.1 |
0.3 |
GO:1900133 |
renin secretion into blood stream(GO:0002001) regulation of renin secretion into blood stream(GO:1900133) |
| 0.1 |
0.3 |
GO:0097401 |
synaptic vesicle lumen acidification(GO:0097401) |
| 0.1 |
0.3 |
GO:0045014 |
detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.1 |
0.3 |
GO:0009946 |
optic vesicle morphogenesis(GO:0003404) proximal/distal axis specification(GO:0009946) neuroblast migration(GO:0097402) |
| 0.1 |
0.4 |
GO:0007113 |
endomitotic cell cycle(GO:0007113) |
| 0.1 |
0.2 |
GO:0070885 |
negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.1 |
0.4 |
GO:1901204 |
positive regulation of guanylate cyclase activity(GO:0031284) regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 0.1 |
0.4 |
GO:0019509 |
L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 |
0.3 |
GO:2000744 |
anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.1 |
0.3 |
GO:0043456 |
regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 |
0.8 |
GO:1904587 |
glycoprotein ERAD pathway(GO:0097466) response to glycoprotein(GO:1904587) |
| 0.1 |
0.3 |
GO:0032513 |
negative regulation of protein phosphatase type 2B activity(GO:0032513) |
| 0.1 |
0.7 |
GO:0033564 |
anterior/posterior axon guidance(GO:0033564) |
| 0.1 |
0.3 |
GO:0019676 |
ammonia assimilation cycle(GO:0019676) |
| 0.1 |
0.4 |
GO:0010288 |
response to lead ion(GO:0010288) |
| 0.1 |
0.2 |
GO:0003100 |
regulation of systemic arterial blood pressure by endothelin(GO:0003100) |
| 0.1 |
0.9 |
GO:0030388 |
fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 |
0.6 |
GO:1900029 |
positive regulation of ruffle assembly(GO:1900029) |
| 0.1 |
0.3 |
GO:0072531 |
pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.1 |
0.3 |
GO:0060266 |
positive regulation of respiratory burst involved in inflammatory response(GO:0060265) negative regulation of respiratory burst involved in inflammatory response(GO:0060266) negative regulation of respiratory burst(GO:0060268) |
| 0.1 |
0.8 |
GO:0010764 |
negative regulation of fibroblast migration(GO:0010764) |
| 0.1 |
0.3 |
GO:0046552 |
eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.1 |
0.7 |
GO:0021894 |
cerebral cortex GABAergic interneuron development(GO:0021894) |
| 0.1 |
0.2 |
GO:1900078 |
positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.1 |
0.4 |
GO:1902856 |
negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.1 |
0.1 |
GO:0032095 |
regulation of response to food(GO:0032095) negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.1 |
0.2 |
GO:0003289 |
atrial septum primum morphogenesis(GO:0003289) |
| 0.1 |
0.3 |
GO:0051866 |
general adaptation syndrome(GO:0051866) |
| 0.1 |
0.4 |
GO:0031033 |
myosin filament organization(GO:0031033) |
| 0.1 |
0.4 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.1 |
0.2 |
GO:1902109 |
negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.1 |
0.9 |
GO:0061302 |
smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 |
0.2 |
GO:0030240 |
skeletal muscle thin filament assembly(GO:0030240) |
| 0.1 |
0.5 |
GO:0036444 |
calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 |
0.4 |
GO:0071672 |
negative regulation of smooth muscle cell chemotaxis(GO:0071672) |
| 0.1 |
0.5 |
GO:0008343 |
adult feeding behavior(GO:0008343) |
| 0.1 |
0.3 |
GO:0090238 |
positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.1 |
0.4 |
GO:0051490 |
negative regulation of filopodium assembly(GO:0051490) |
| 0.1 |
0.1 |
GO:0032227 |
negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.1 |
0.2 |
GO:0072070 |
establishment or maintenance of polarity of embryonic epithelium(GO:0016332) loop of Henle development(GO:0072070) metanephric loop of Henle development(GO:0072236) |
| 0.1 |
0.2 |
GO:0035864 |
response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.1 |
1.0 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 |
0.3 |
GO:2000383 |
regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.1 |
0.3 |
GO:0090038 |
negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 |
0.3 |
GO:0046600 |
negative regulation of centriole replication(GO:0046600) |
| 0.1 |
0.2 |
GO:0070212 |
protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 |
0.8 |
GO:2000653 |
regulation of genetic imprinting(GO:2000653) |
| 0.1 |
0.3 |
GO:0021564 |
glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 0.1 |
0.3 |
GO:0070829 |
heterochromatin maintenance(GO:0070829) |
| 0.1 |
0.3 |
GO:0097527 |
necroptotic signaling pathway(GO:0097527) |
| 0.1 |
0.1 |
GO:0051599 |
response to hydrostatic pressure(GO:0051599) |
| 0.1 |
0.3 |
GO:1903298 |
regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) |
| 0.1 |
0.1 |
GO:1902969 |
mitotic DNA replication(GO:1902969) |
| 0.1 |
0.1 |
GO:0007341 |
penetration of zona pellucida(GO:0007341) |
| 0.1 |
0.3 |
GO:0043985 |
histone H4-R3 methylation(GO:0043985) |
| 0.1 |
1.4 |
GO:0060632 |
regulation of microtubule-based movement(GO:0060632) |
| 0.1 |
0.3 |
GO:0002337 |
B-1a B cell differentiation(GO:0002337) |
| 0.1 |
0.1 |
GO:2000813 |
negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.1 |
0.4 |
GO:0006527 |
arginine catabolic process(GO:0006527) |
| 0.1 |
0.3 |
GO:0048319 |
axial mesoderm morphogenesis(GO:0048319) |
| 0.1 |
0.3 |
GO:0018002 |
N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.1 |
0.2 |
GO:1900533 |
medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.1 |
0.2 |
GO:0015786 |
UDP-glucose transport(GO:0015786) |
| 0.1 |
0.2 |
GO:0097167 |
circadian regulation of translation(GO:0097167) |
| 0.1 |
0.3 |
GO:0006741 |
NADP biosynthetic process(GO:0006741) |
| 0.1 |
0.3 |
GO:0051697 |
protein delipidation(GO:0051697) |
| 0.1 |
0.1 |
GO:0071550 |
death-inducing signaling complex assembly(GO:0071550) |
| 0.1 |
0.2 |
GO:1904529 |
regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.1 |
0.5 |
GO:0031642 |
negative regulation of myelination(GO:0031642) |
| 0.1 |
0.2 |
GO:0045903 |
positive regulation of translational fidelity(GO:0045903) |
| 0.1 |
0.1 |
GO:0043585 |
nose morphogenesis(GO:0043585) |
| 0.1 |
0.4 |
GO:0046541 |
saliva secretion(GO:0046541) |
| 0.1 |
0.1 |
GO:0060120 |
auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.1 |
0.2 |
GO:0071492 |
cellular response to UV-A(GO:0071492) |
| 0.1 |
0.2 |
GO:0035990 |
tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.1 |
0.5 |
GO:0046036 |
CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.1 |
0.3 |
GO:0015803 |
branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 |
0.1 |
GO:2000297 |
negative regulation of synapse maturation(GO:2000297) |
| 0.1 |
1.0 |
GO:0051016 |
barbed-end actin filament capping(GO:0051016) |
| 0.1 |
0.2 |
GO:0010637 |
negative regulation of mitochondrial fusion(GO:0010637) |
| 0.1 |
0.3 |
GO:0019732 |
antifungal humoral response(GO:0019732) |
| 0.1 |
0.2 |
GO:0007227 |
signal transduction downstream of smoothened(GO:0007227) positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.1 |
0.2 |
GO:0071105 |
response to interleukin-9(GO:0071104) response to interleukin-11(GO:0071105) |
| 0.1 |
1.3 |
GO:0045663 |
positive regulation of myoblast differentiation(GO:0045663) |
| 0.1 |
0.1 |
GO:0016340 |
calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.1 |
0.5 |
GO:0006012 |
galactose metabolic process(GO:0006012) |
| 0.1 |
0.7 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 |
0.2 |
GO:0009106 |
lipoate metabolic process(GO:0009106) |
| 0.1 |
0.9 |
GO:0031115 |
negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 |
0.2 |
GO:0006407 |
rRNA export from nucleus(GO:0006407) |
| 0.1 |
0.2 |
GO:1901492 |
positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 |
0.1 |
GO:0014051 |
gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 |
0.1 |
GO:0030208 |
dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 |
0.1 |
GO:0046101 |
hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 |
0.1 |
GO:1902219 |
regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.0 |
0.1 |
GO:0061738 |
abscission(GO:0009838) late endosomal microautophagy(GO:0061738) positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 |
0.7 |
GO:0070986 |
left/right axis specification(GO:0070986) |
| 0.0 |
0.4 |
GO:0031424 |
keratinization(GO:0031424) |
| 0.0 |
0.4 |
GO:0032625 |
interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 |
0.2 |
GO:0060155 |
platelet dense granule organization(GO:0060155) |
| 0.0 |
0.2 |
GO:0070934 |
CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 |
0.0 |
GO:0009597 |
detection of virus(GO:0009597) |
| 0.0 |
0.1 |
GO:1900623 |
regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 |
0.2 |
GO:0006369 |
termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 |
0.7 |
GO:0008535 |
respiratory chain complex IV assembly(GO:0008535) |
| 0.0 |
0.0 |
GO:0045794 |
negative regulation of cell volume(GO:0045794) |
| 0.0 |
0.2 |
GO:0014046 |
dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 |
0.1 |
GO:0016056 |
rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 |
0.1 |
GO:0031958 |
corticosteroid receptor signaling pathway(GO:0031958) |
| 0.0 |
0.1 |
GO:0006672 |
ceramide metabolic process(GO:0006672) |
| 0.0 |
0.2 |
GO:0032472 |
Golgi calcium ion transport(GO:0032472) |
| 0.0 |
0.2 |
GO:1903265 |
positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 |
0.2 |
GO:0010961 |
cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 |
0.1 |
GO:0008626 |
granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 |
0.2 |
GO:0018125 |
peptidyl-cysteine methylation(GO:0018125) |
| 0.0 |
0.2 |
GO:0060158 |
phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 |
0.4 |
GO:0044387 |
negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.0 |
0.2 |
GO:0031509 |
telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.0 |
0.3 |
GO:0005981 |
regulation of glycogen catabolic process(GO:0005981) |
| 0.0 |
0.3 |
GO:2000124 |
regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 |
0.3 |
GO:0038028 |
insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 |
0.1 |
GO:0034776 |
response to histamine(GO:0034776) cellular response to histamine(GO:0071420) |
| 0.0 |
0.1 |
GO:0071544 |
diphosphoinositol polyphosphate metabolic process(GO:0071543) diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.0 |
0.4 |
GO:0035970 |
peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 |
0.1 |
GO:0006931 |
substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 |
0.1 |
GO:0019043 |
establishment of viral latency(GO:0019043) |
| 0.0 |
0.2 |
GO:0035726 |
common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 |
0.2 |
GO:0046543 |
development of secondary female sexual characteristics(GO:0046543) |
| 0.0 |
0.2 |
GO:0030576 |
Cajal body organization(GO:0030576) |
| 0.0 |
0.1 |
GO:0018298 |
protein-chromophore linkage(GO:0018298) |
| 0.0 |
0.2 |
GO:0070141 |
response to UV-A(GO:0070141) |
| 0.0 |
0.1 |
GO:0010940 |
positive regulation of necrotic cell death(GO:0010940) |
| 0.0 |
0.1 |
GO:0042271 |
susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 |
0.4 |
GO:1990173 |
protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.0 |
0.2 |
GO:1902309 |
negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 |
0.1 |
GO:0000715 |
nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 |
0.2 |
GO:0090205 |
positive regulation of cholesterol biosynthetic process(GO:0045542) positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.0 |
0.7 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.0 |
0.3 |
GO:0043951 |
negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 |
0.2 |
GO:0071494 |
cellular response to UV-C(GO:0071494) |
| 0.0 |
0.2 |
GO:1901385 |
regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 |
0.2 |
GO:0006189 |
'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 |
0.2 |
GO:0032382 |
positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) positive regulation of cholesterol import(GO:1904109) positive regulation of sterol import(GO:2000911) |
| 0.0 |
0.1 |
GO:0018214 |
peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 |
0.3 |
GO:0010225 |
response to UV-C(GO:0010225) |
| 0.0 |
0.2 |
GO:0002536 |
respiratory burst involved in inflammatory response(GO:0002536) |
| 0.0 |
0.6 |
GO:0006107 |
oxaloacetate metabolic process(GO:0006107) |
| 0.0 |
0.2 |
GO:0070164 |
negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 |
0.2 |
GO:2000672 |
negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 |
0.1 |
GO:0035672 |
transepithelial chloride transport(GO:0030321) oligopeptide transmembrane transport(GO:0035672) |
| 0.0 |
0.3 |
GO:0018401 |
peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 |
0.2 |
GO:0046208 |
spermine catabolic process(GO:0046208) |
| 0.0 |
0.2 |
GO:0003420 |
regulation of growth plate cartilage chondrocyte proliferation(GO:0003420) |
| 0.0 |
0.1 |
GO:0036258 |
multivesicular body assembly(GO:0036258) |
| 0.0 |
0.5 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 |
0.1 |
GO:0060684 |
epithelial-mesenchymal cell signaling(GO:0060684) |
| 0.0 |
0.1 |
GO:0002035 |
brain renin-angiotensin system(GO:0002035) |
| 0.0 |
0.6 |
GO:0042744 |
hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 |
0.5 |
GO:0071257 |
cellular response to electrical stimulus(GO:0071257) |
| 0.0 |
0.8 |
GO:0007020 |
microtubule nucleation(GO:0007020) |
| 0.0 |
0.2 |
GO:0015015 |
heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 |
0.1 |
GO:2000507 |
positive regulation of energy homeostasis(GO:2000507) |
| 0.0 |
0.1 |
GO:0070973 |
protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 |
0.4 |
GO:0019262 |
N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 |
0.2 |
GO:0060040 |
retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 |
0.1 |
GO:0000733 |
DNA strand renaturation(GO:0000733) |
| 0.0 |
0.2 |
GO:1901096 |
regulation of autophagosome maturation(GO:1901096) |
| 0.0 |
0.1 |
GO:1903966 |
monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 |
0.3 |
GO:0032464 |
positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 |
0.2 |
GO:1902715 |
positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.0 |
0.2 |
GO:0021993 |
initiation of neural tube closure(GO:0021993) |
| 0.0 |
0.9 |
GO:0018345 |
protein palmitoylation(GO:0018345) |
| 0.0 |
0.1 |
GO:0002302 |
CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.0 |
0.1 |
GO:1900748 |
positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 |
0.1 |
GO:0006867 |
asparagine transport(GO:0006867) glutamine transport(GO:0006868) |
| 0.0 |
0.2 |
GO:0006344 |
maintenance of chromatin silencing(GO:0006344) |
| 0.0 |
0.1 |
GO:0021577 |
hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) lateral motor column neuron migration(GO:0097477) |
| 0.0 |
0.1 |
GO:0002014 |
vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.0 |
0.1 |
GO:0048170 |
positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 |
0.3 |
GO:0036289 |
peptidyl-serine autophosphorylation(GO:0036289) |
| 0.0 |
0.0 |
GO:0019074 |
viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 |
0.3 |
GO:0097688 |
AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.0 |
0.4 |
GO:0006995 |
cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 |
0.3 |
GO:0010718 |
positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 |
0.3 |
GO:0045607 |
regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 |
0.2 |
GO:0009145 |
ATP biosynthetic process(GO:0006754) purine nucleoside triphosphate biosynthetic process(GO:0009145) ribonucleoside triphosphate biosynthetic process(GO:0009201) purine ribonucleoside triphosphate biosynthetic process(GO:0009206) |
| 0.0 |
0.1 |
GO:1902268 |
negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 |
0.1 |
GO:0071895 |
odontoblast differentiation(GO:0071895) |
| 0.0 |
0.4 |
GO:0007076 |
mitotic chromosome condensation(GO:0007076) |
| 0.0 |
0.1 |
GO:0019355 |
nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 |
0.1 |
GO:0060598 |
dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.0 |
0.3 |
GO:0046599 |
regulation of centriole replication(GO:0046599) |
| 0.0 |
0.2 |
GO:0001842 |
neural fold formation(GO:0001842) |
| 0.0 |
0.3 |
GO:0016198 |
axon choice point recognition(GO:0016198) |
| 0.0 |
0.1 |
GO:0051835 |
positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 |
0.4 |
GO:0007252 |
I-kappaB phosphorylation(GO:0007252) |
| 0.0 |
0.1 |
GO:1902277 |
negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.0 |
0.0 |
GO:0061153 |
trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 |
0.2 |
GO:0090164 |
asymmetric Golgi ribbon formation(GO:0090164) |
| 0.0 |
0.1 |
GO:0061526 |
acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) acetylcholine secretion(GO:0061526) |
| 0.0 |
0.4 |
GO:0060052 |
neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 |
0.2 |
GO:0000160 |
phosphorelay signal transduction system(GO:0000160) |
| 0.0 |
0.1 |
GO:0071985 |
multivesicular body sorting pathway(GO:0071985) |
| 0.0 |
0.2 |
GO:0008355 |
olfactory learning(GO:0008355) |
| 0.0 |
0.2 |
GO:1903204 |
negative regulation of oxidative stress-induced neuron death(GO:1903204) |
| 0.0 |
0.3 |
GO:0090036 |
regulation of protein kinase C signaling(GO:0090036) |
| 0.0 |
0.5 |
GO:0007530 |
sex determination(GO:0007530) |
| 0.0 |
0.5 |
GO:0007263 |
nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 |
0.1 |
GO:2001015 |
negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.0 |
0.1 |
GO:1900242 |
regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.0 |
0.1 |
GO:0072592 |
oxygen metabolic process(GO:0072592) |
| 0.0 |
0.1 |
GO:0014832 |
urinary bladder smooth muscle contraction(GO:0014832) |
| 0.0 |
0.1 |
GO:0046340 |
diacylglycerol catabolic process(GO:0046340) |
| 0.0 |
0.2 |
GO:0031639 |
plasminogen activation(GO:0031639) |
| 0.0 |
0.2 |
GO:0048227 |
plasma membrane to endosome transport(GO:0048227) |
| 0.0 |
0.2 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.0 |
0.3 |
GO:2000480 |
negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 |
0.1 |
GO:0044268 |
multicellular organismal protein metabolic process(GO:0044268) |
| 0.0 |
0.1 |
GO:0035878 |
nail development(GO:0035878) |
| 0.0 |
0.2 |
GO:0042699 |
follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 |
0.1 |
GO:0006382 |
adenosine to inosine editing(GO:0006382) |
| 0.0 |
0.2 |
GO:0060837 |
blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 |
0.2 |
GO:0030579 |
ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 |
0.2 |
GO:0050765 |
negative regulation of phagocytosis(GO:0050765) |
| 0.0 |
0.1 |
GO:1900365 |
positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 |
0.2 |
GO:0090342 |
regulation of cell aging(GO:0090342) regulation of cellular senescence(GO:2000772) |
| 0.0 |
0.3 |
GO:0070445 |
oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 |
0.0 |
GO:0046888 |
negative regulation of hormone secretion(GO:0046888) |
| 0.0 |
0.3 |
GO:2000009 |
negative regulation of protein localization to cell surface(GO:2000009) |
| 0.0 |
0.7 |
GO:0095500 |
acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 |
0.6 |
GO:0007205 |
protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 |
0.3 |
GO:0031274 |
positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 |
0.0 |
GO:2000642 |
negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.0 |
0.2 |
GO:0034587 |
piRNA metabolic process(GO:0034587) |
| 0.0 |
0.1 |
GO:0072643 |
interferon-gamma secretion(GO:0072643) |
| 0.0 |
1.0 |
GO:0006890 |
retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 |
0.1 |
GO:0019287 |
isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.0 |
0.1 |
GO:0090370 |
negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 |
0.2 |
GO:0097067 |
cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 |
0.1 |
GO:0098903 |
regulation of membrane repolarization during action potential(GO:0098903) |
| 0.0 |
0.2 |
GO:0007130 |
synaptonemal complex assembly(GO:0007130) |
| 0.0 |
0.2 |
GO:0031119 |
tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 |
0.1 |
GO:2001137 |
positive regulation of endocytic recycling(GO:2001137) |
| 0.0 |
0.1 |
GO:0002756 |
MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.0 |
0.3 |
GO:0051930 |
regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.0 |
0.3 |
GO:0030150 |
protein import into mitochondrial matrix(GO:0030150) |
| 0.0 |
0.2 |
GO:0051152 |
positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 |
0.1 |
GO:0002159 |
desmosome assembly(GO:0002159) |
| 0.0 |
0.1 |
GO:0090273 |
regulation of somatostatin secretion(GO:0090273) |
| 0.0 |
0.1 |
GO:0045054 |
constitutive secretory pathway(GO:0045054) |
| 0.0 |
0.1 |
GO:1903300 |
negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 |
0.1 |
GO:0061144 |
alveolar secondary septum development(GO:0061144) |
| 0.0 |
0.1 |
GO:1904694 |
negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 |
0.1 |
GO:0045003 |
DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 |
0.4 |
GO:0060065 |
uterus development(GO:0060065) |
| 0.0 |
0.2 |
GO:0006003 |
fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 |
0.1 |
GO:0006669 |
sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 |
0.1 |
GO:0050909 |
sensory perception of taste(GO:0050909) |
| 0.0 |
0.2 |
GO:0032525 |
somite rostral/caudal axis specification(GO:0032525) |
| 0.0 |
0.1 |
GO:2001045 |
negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 |
0.1 |
GO:0034638 |
phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 |
0.1 |
GO:0035461 |
vitamin transmembrane transport(GO:0035461) |
| 0.0 |
0.0 |
GO:0032847 |
regulation of cellular pH reduction(GO:0032847) |
| 0.0 |
0.3 |
GO:0023041 |
neuronal signal transduction(GO:0023041) |
| 0.0 |
0.3 |
GO:0090244 |
Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.0 |
0.1 |
GO:0048680 |
positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 |
0.1 |
GO:1903715 |
regulation of aerobic respiration(GO:1903715) |
| 0.0 |
0.1 |
GO:0034035 |
purine ribonucleoside bisphosphate metabolic process(GO:0034035) |
| 0.0 |
0.0 |
GO:0044332 |
Wnt signaling pathway involved in dorsal/ventral axis specification(GO:0044332) |
| 0.0 |
0.1 |
GO:0098838 |
reduced folate transmembrane transport(GO:0098838) |
| 0.0 |
0.2 |
GO:0039703 |
viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 |
0.3 |
GO:0010388 |
protein deneddylation(GO:0000338) cullin deneddylation(GO:0010388) |
| 0.0 |
0.1 |
GO:0000098 |
sulfur amino acid catabolic process(GO:0000098) |
| 0.0 |
0.2 |
GO:0042659 |
regulation of cell fate specification(GO:0042659) |
| 0.0 |
0.1 |
GO:0007253 |
cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 |
0.1 |
GO:2000301 |
negative regulation of synaptic vesicle transport(GO:1902804) negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 |
0.1 |
GO:0009182 |
purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) dGDP metabolic process(GO:0046066) GDP metabolic process(GO:0046710) guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 |
0.0 |
GO:0035434 |
copper ion transmembrane transport(GO:0035434) |
| 0.0 |
0.3 |
GO:0002115 |
store-operated calcium entry(GO:0002115) |
| 0.0 |
0.1 |
GO:0006450 |
regulation of translational fidelity(GO:0006450) |
| 0.0 |
0.1 |
GO:0071236 |
cellular response to antibiotic(GO:0071236) |
| 0.0 |
1.3 |
GO:0032543 |
mitochondrial translation(GO:0032543) |
| 0.0 |
0.3 |
GO:0014047 |
glutamate secretion(GO:0014047) |
| 0.0 |
0.1 |
GO:2001012 |
mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.0 |
0.1 |
GO:0060231 |
mesenchymal to epithelial transition(GO:0060231) |
| 0.0 |
0.2 |
GO:2000254 |
regulation of male germ cell proliferation(GO:2000254) |
| 0.0 |
0.5 |
GO:0033622 |
integrin activation(GO:0033622) |
| 0.0 |
0.1 |
GO:0048069 |
eye pigmentation(GO:0048069) |
| 0.0 |
0.5 |
GO:0043949 |
regulation of cAMP-mediated signaling(GO:0043949) |
| 0.0 |
0.2 |
GO:0019243 |
methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 |
0.3 |
GO:0045116 |
protein neddylation(GO:0045116) |
| 0.0 |
0.2 |
GO:0010992 |
ubiquitin homeostasis(GO:0010992) |
| 0.0 |
0.0 |
GO:2001245 |
regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 |
0.1 |
GO:0006097 |
glyoxylate cycle(GO:0006097) |
| 0.0 |
0.2 |
GO:0051386 |
regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.0 |
0.1 |
GO:0048012 |
hepatocyte growth factor receptor signaling pathway(GO:0048012) |
| 0.0 |
0.1 |
GO:0033762 |
response to glucagon(GO:0033762) |
| 0.0 |
0.1 |
GO:0097284 |
hepatocyte apoptotic process(GO:0097284) |
| 0.0 |
0.1 |
GO:2000172 |
regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.0 |
0.8 |
GO:0032233 |
positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 |
0.1 |
GO:0016264 |
gap junction assembly(GO:0016264) |
| 0.0 |
0.1 |
GO:0010359 |
regulation of anion channel activity(GO:0010359) |
| 0.0 |
0.2 |
GO:1901299 |
negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.0 |
0.1 |
GO:2000483 |
negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 |
0.1 |
GO:0003433 |
chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.0 |
0.1 |
GO:1902498 |
regulation of protein autoubiquitination(GO:1902498) |
| 0.0 |
0.2 |
GO:0008054 |
negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) |
| 0.0 |
0.1 |
GO:0036438 |
maintenance of lens transparency(GO:0036438) |
| 0.0 |
0.3 |
GO:0030252 |
growth hormone secretion(GO:0030252) |
| 0.0 |
0.5 |
GO:0016082 |
synaptic vesicle priming(GO:0016082) |
| 0.0 |
0.1 |
GO:1901301 |
regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 |
0.1 |
GO:0070682 |
proteasome regulatory particle assembly(GO:0070682) |
| 0.0 |
0.3 |
GO:0035563 |
positive regulation of chromatin binding(GO:0035563) |
| 0.0 |
0.2 |
GO:0048096 |
chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 |
0.1 |
GO:0072429 |
response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 |
0.1 |
GO:0070458 |
detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.0 |
0.1 |
GO:0006044 |
N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 |
0.1 |
GO:1904263 |
positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 |
0.9 |
GO:0009060 |
aerobic respiration(GO:0009060) |
| 0.0 |
0.2 |
GO:0005980 |
polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 |
0.1 |
GO:1902902 |
negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 |
0.3 |
GO:2000060 |
positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.0 |
0.2 |
GO:0051205 |
protein insertion into membrane(GO:0051205) |
| 0.0 |
0.5 |
GO:0070979 |
protein K11-linked ubiquitination(GO:0070979) |
| 0.0 |
0.1 |
GO:0018364 |
peptidyl-glutamine methylation(GO:0018364) |
| 0.0 |
0.2 |
GO:0031507 |
heterochromatin assembly(GO:0031507) |
| 0.0 |
0.0 |
GO:0007603 |
phototransduction, visible light(GO:0007603) |
| 0.0 |
0.1 |
GO:0010917 |
negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.0 |
0.4 |
GO:0090280 |
positive regulation of calcium ion import(GO:0090280) |
| 0.0 |
0.2 |
GO:0010667 |
negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 |
0.1 |
GO:0045041 |
protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 |
0.1 |
GO:0010572 |
positive regulation of platelet activation(GO:0010572) |
| 0.0 |
0.1 |
GO:2000491 |
positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 |
0.1 |
GO:0007343 |
egg activation(GO:0007343) |
| 0.0 |
0.1 |
GO:0090309 |
positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 |
0.1 |
GO:0070428 |
nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.0 |
0.1 |
GO:0035845 |
photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 |
0.1 |
GO:0009188 |
ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.0 |
0.2 |
GO:0043248 |
proteasome assembly(GO:0043248) |
| 0.0 |
0.1 |
GO:0090394 |
negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 0.0 |
0.4 |
GO:0060706 |
cell differentiation involved in embryonic placenta development(GO:0060706) |
| 0.0 |
0.1 |
GO:0070584 |
mitochondrion morphogenesis(GO:0070584) |
| 0.0 |
0.0 |
GO:0032898 |
neurotrophin production(GO:0032898) |
| 0.0 |
0.8 |
GO:0007019 |
microtubule depolymerization(GO:0007019) |
| 0.0 |
0.2 |
GO:1903333 |
negative regulation of protein folding(GO:1903333) |
| 0.0 |
0.2 |
GO:0035721 |
intraciliary retrograde transport(GO:0035721) |
| 0.0 |
0.1 |
GO:0015819 |
lysine transport(GO:0015819) |
| 0.0 |
0.5 |
GO:0046513 |
ceramide biosynthetic process(GO:0046513) |
| 0.0 |
0.1 |
GO:0046085 |
adenosine metabolic process(GO:0046085) |
| 0.0 |
0.1 |
GO:0002002 |
regulation of systemic arterial blood pressure by circulatory renin-angiotensin(GO:0001991) regulation of angiotensin levels in blood(GO:0002002) |
| 0.0 |
0.2 |
GO:0003334 |
keratinocyte development(GO:0003334) |
| 0.0 |
0.0 |
GO:0038003 |
opioid receptor signaling pathway(GO:0038003) regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.0 |
0.1 |
GO:0035871 |
protein K11-linked deubiquitination(GO:0035871) |
| 0.0 |
0.1 |
GO:0014029 |
neural crest formation(GO:0014029) |
| 0.0 |
0.1 |
GO:0075525 |
viral translational termination-reinitiation(GO:0075525) |
| 0.0 |
0.0 |
GO:0021759 |
globus pallidus development(GO:0021759) |
| 0.0 |
0.1 |
GO:0006776 |
vitamin A metabolic process(GO:0006776) |
| 0.0 |
0.1 |
GO:0035428 |
hexose transmembrane transport(GO:0035428) |
| 0.0 |
0.2 |
GO:0045672 |
positive regulation of osteoclast differentiation(GO:0045672) |
| 0.0 |
0.1 |
GO:2000727 |
positive regulation of cardiac muscle cell differentiation(GO:2000727) |
| 0.0 |
0.2 |
GO:0000060 |
protein import into nucleus, translocation(GO:0000060) |
| 0.0 |
0.1 |
GO:1901020 |
negative regulation of calcium ion transmembrane transporter activity(GO:1901020) |
| 0.0 |
0.2 |
GO:0035269 |
protein O-linked mannosylation(GO:0035269) |
| 0.0 |
0.1 |
GO:0000103 |
sulfate assimilation(GO:0000103) |
| 0.0 |
0.1 |
GO:0043314 |
negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) negative regulation of neutrophil activation(GO:1902564) |
| 0.0 |
0.3 |
GO:0048026 |
positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 |
0.1 |
GO:0032808 |
lacrimal gland development(GO:0032808) |
| 0.0 |
0.1 |
GO:0010815 |
bradykinin catabolic process(GO:0010815) |
| 0.0 |
0.1 |
GO:0042346 |
positive regulation of NF-kappaB import into nucleus(GO:0042346) |
| 0.0 |
0.1 |
GO:1901475 |
pyruvate transmembrane transport(GO:1901475) |
| 0.0 |
0.0 |
GO:1903599 |
positive regulation of mitophagy(GO:1903599) |
| 0.0 |
0.0 |
GO:0018199 |
peptidyl-glutamine modification(GO:0018199) |
| 0.0 |
0.2 |
GO:0001731 |
formation of translation preinitiation complex(GO:0001731) |
| 0.0 |
0.2 |
GO:0045947 |
negative regulation of translational initiation(GO:0045947) |
| 0.0 |
0.3 |
GO:0034260 |
negative regulation of GTPase activity(GO:0034260) |
| 0.0 |
0.5 |
GO:0018279 |
peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 |
0.1 |
GO:0018065 |
protein-cofactor linkage(GO:0018065) |
| 0.0 |
0.0 |
GO:0000821 |
regulation of arginine metabolic process(GO:0000821) |
| 0.0 |
0.0 |
GO:0039536 |
negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 |
0.1 |
GO:0035887 |
aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 |
0.1 |
GO:0045742 |
positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.0 |
0.1 |
GO:0035025 |
positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 |
0.1 |
GO:0006123 |
mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 |
0.2 |
GO:0051560 |
mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 |
0.2 |
GO:0048025 |
negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 |
0.1 |
GO:0042866 |
pyruvate biosynthetic process(GO:0042866) |
| 0.0 |
0.1 |
GO:0060633 |
negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 |
0.1 |
GO:0023021 |
termination of signal transduction(GO:0023021) |
| 0.0 |
0.1 |
GO:0035609 |
C-terminal protein deglutamylation(GO:0035609) |
| 0.0 |
0.1 |
GO:0042167 |
heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 |
0.1 |
GO:0007000 |
nucleolus organization(GO:0007000) |
| 0.0 |
0.1 |
GO:0007250 |
activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 |
0.0 |
GO:0071929 |
alpha-tubulin acetylation(GO:0071929) positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.0 |
0.1 |
GO:0042711 |
maternal behavior(GO:0042711) |
| 0.0 |
0.1 |
GO:0035066 |
positive regulation of histone acetylation(GO:0035066) |
| 0.0 |
0.1 |
GO:1904217 |
regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 |
0.1 |
GO:2000324 |
positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 |
0.5 |
GO:0001825 |
blastocyst formation(GO:0001825) |
| 0.0 |
0.1 |
GO:0051481 |
negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.0 |
0.1 |
GO:0018095 |
protein polyglutamylation(GO:0018095) |
| 0.0 |
0.0 |
GO:1902229 |
regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902229) negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 |
0.0 |
GO:0060217 |
positive regulation of chromatin assembly or disassembly(GO:0045799) hemangioblast cell differentiation(GO:0060217) |
| 0.0 |
0.2 |
GO:0035235 |
ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 |
0.2 |
GO:0016558 |
protein import into peroxisome matrix(GO:0016558) |
| 0.0 |
0.0 |
GO:0042373 |
vitamin K metabolic process(GO:0042373) |
| 0.0 |
0.1 |
GO:0007220 |
Notch receptor processing(GO:0007220) |
| 0.0 |
0.1 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.0 |
0.3 |
GO:2000785 |
regulation of autophagosome assembly(GO:2000785) |
| 0.0 |
0.1 |
GO:0032462 |
regulation of protein homooligomerization(GO:0032462) |
| 0.0 |
0.0 |
GO:0072718 |
response to cisplatin(GO:0072718) |
| 0.0 |
0.1 |
GO:0090503 |
RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 |
0.0 |
GO:2000195 |
negative regulation of female gonad development(GO:2000195) |
| 0.0 |
0.1 |
GO:0097352 |
autophagosome maturation(GO:0097352) |
| 0.0 |
0.2 |
GO:0045943 |
positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 |
0.0 |
GO:0006103 |
2-oxoglutarate metabolic process(GO:0006103) |
| 0.0 |
0.1 |
GO:0000185 |
activation of MAPKKK activity(GO:0000185) |
| 0.0 |
0.0 |
GO:0043490 |
malate-aspartate shuttle(GO:0043490) |
| 0.0 |
0.2 |
GO:0030213 |
hyaluronan biosynthetic process(GO:0030213) |
| 0.0 |
0.5 |
GO:0016925 |
protein sumoylation(GO:0016925) |
| 0.0 |
0.1 |
GO:0042059 |
negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.0 |
0.1 |
GO:0071472 |
cellular response to salt stress(GO:0071472) |
| 0.0 |
0.1 |
GO:0035646 |
endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.0 |
0.1 |
GO:0052646 |
alditol phosphate metabolic process(GO:0052646) |
| 0.0 |
0.2 |
GO:0090201 |
negative regulation of release of cytochrome c from mitochondria(GO:0090201) |
| 0.0 |
0.0 |
GO:0090399 |
replicative senescence(GO:0090399) |
| 0.0 |
0.2 |
GO:0009072 |
aromatic amino acid family metabolic process(GO:0009072) |
| 0.0 |
0.6 |
GO:0060976 |
coronary vasculature development(GO:0060976) |
| 0.0 |
0.1 |
GO:0045830 |
positive regulation of isotype switching(GO:0045830) |
| 0.0 |
0.0 |
GO:0051534 |
negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 |
0.0 |
GO:0051725 |
protein de-ADP-ribosylation(GO:0051725) |
| 0.0 |
0.1 |
GO:0006654 |
phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 |
0.0 |
GO:0048386 |
positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 |
0.1 |
GO:0021681 |
cerebellar granular layer development(GO:0021681) |
| 0.0 |
0.3 |
GO:0006414 |
translational elongation(GO:0006414) |
| 0.0 |
0.2 |
GO:0071480 |
cellular response to gamma radiation(GO:0071480) |
| 0.0 |
0.0 |
GO:0046060 |
dATP metabolic process(GO:0046060) dATP catabolic process(GO:0046061) |
| 0.0 |
0.1 |
GO:0006020 |
inositol metabolic process(GO:0006020) |
| 0.0 |
0.1 |
GO:0007218 |
neuropeptide signaling pathway(GO:0007218) |
| 0.0 |
0.1 |
GO:0007616 |
long-term memory(GO:0007616) |
| 0.0 |
0.0 |
GO:1902047 |
polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) |
| 0.0 |
0.0 |
GO:2000809 |
positive regulation of synaptic vesicle clustering(GO:2000809) |