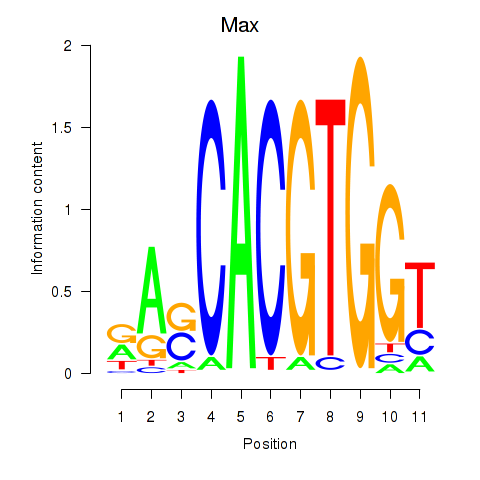
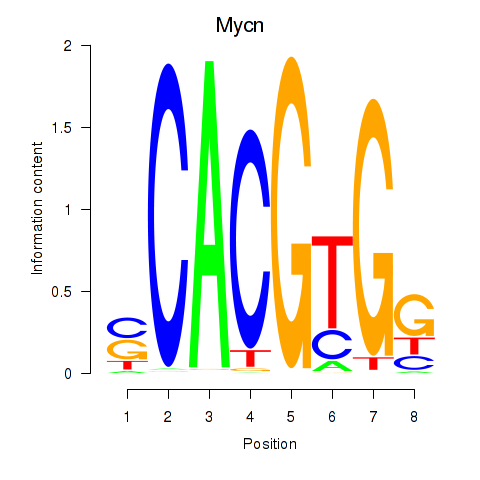
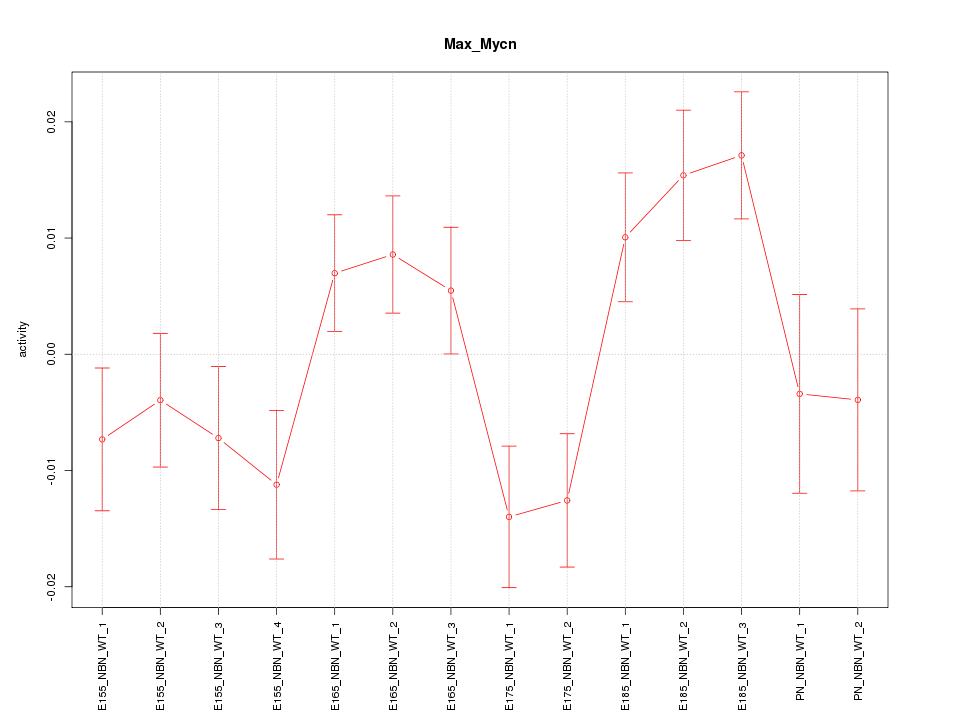
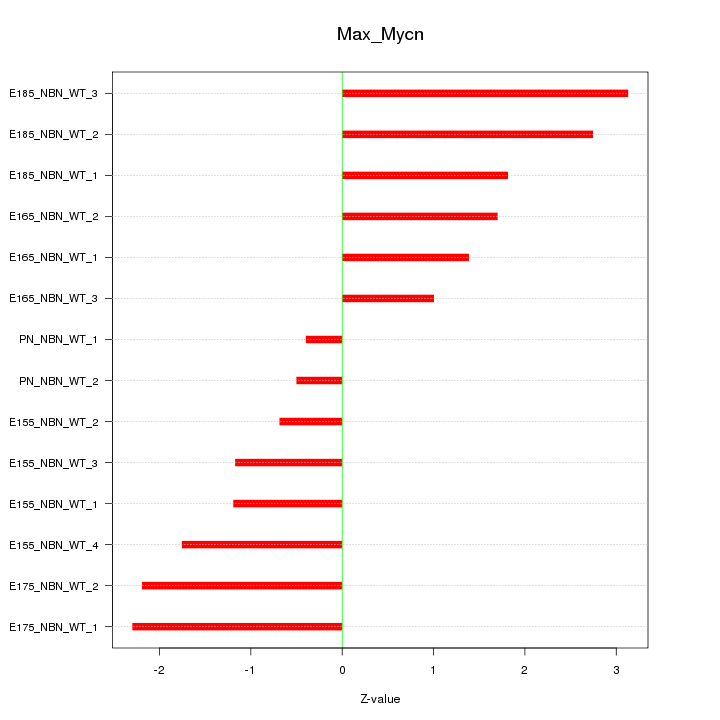
| 0.8 |
3.4 |
GO:0010288 |
response to lead ion(GO:0010288) |
| 0.8 |
4.7 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.6 |
1.7 |
GO:0015712 |
hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.6 |
2.2 |
GO:0060266 |
positive regulation of respiratory burst involved in inflammatory response(GO:0060265) negative regulation of respiratory burst involved in inflammatory response(GO:0060266) negative regulation of respiratory burst(GO:0060268) |
| 0.5 |
1.5 |
GO:0009446 |
putrescine biosynthetic process(GO:0009446) |
| 0.5 |
1.5 |
GO:0019343 |
cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) |
| 0.5 |
1.4 |
GO:2000852 |
regulation of corticosterone secretion(GO:2000852) |
| 0.5 |
1.4 |
GO:0019043 |
establishment of viral latency(GO:0019043) |
| 0.5 |
1.4 |
GO:0036166 |
phenotypic switching(GO:0036166) |
| 0.5 |
2.3 |
GO:0043985 |
histone H4-R3 methylation(GO:0043985) |
| 0.4 |
2.5 |
GO:0033600 |
negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.4 |
1.1 |
GO:0008295 |
spermidine biosynthetic process(GO:0008295) |
| 0.4 |
1.1 |
GO:0006059 |
hexitol metabolic process(GO:0006059) |
| 0.4 |
2.9 |
GO:0000050 |
urea cycle(GO:0000050) urea metabolic process(GO:0019627) |
| 0.4 |
1.1 |
GO:0070476 |
rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.3 |
1.4 |
GO:0071051 |
polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.3 |
1.0 |
GO:0045041 |
protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.3 |
0.6 |
GO:0072223 |
metanephric glomerular mesangium development(GO:0072223) metanephric glomerular mesangial cell proliferation involved in metanephros development(GO:0072262) |
| 0.3 |
1.3 |
GO:1902263 |
apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.3 |
2.2 |
GO:0051503 |
adenine nucleotide transport(GO:0051503) |
| 0.3 |
0.9 |
GO:0007386 |
compartment pattern specification(GO:0007386) |
| 0.3 |
1.2 |
GO:0002949 |
tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.3 |
1.5 |
GO:0018002 |
N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.3 |
1.1 |
GO:0072197 |
ureter morphogenesis(GO:0072197) |
| 0.3 |
1.4 |
GO:0045716 |
positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.3 |
1.1 |
GO:0048819 |
regulation of hair follicle maturation(GO:0048819) regulation of catagen(GO:0051794) |
| 0.3 |
1.1 |
GO:0006407 |
rRNA export from nucleus(GO:0006407) |
| 0.3 |
0.8 |
GO:0000720 |
pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.3 |
0.3 |
GO:1901536 |
regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) |
| 0.3 |
1.1 |
GO:2000048 |
negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.3 |
1.0 |
GO:0045903 |
positive regulation of translational fidelity(GO:0045903) |
| 0.2 |
0.7 |
GO:0003195 |
tricuspid valve development(GO:0003175) tricuspid valve morphogenesis(GO:0003186) tricuspid valve formation(GO:0003195) |
| 0.2 |
1.2 |
GO:2000255 |
negative regulation of male germ cell proliferation(GO:2000255) |
| 0.2 |
0.7 |
GO:0060785 |
regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.2 |
1.9 |
GO:0031666 |
positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.2 |
1.6 |
GO:0072321 |
chaperone-mediated protein transport(GO:0072321) |
| 0.2 |
1.2 |
GO:0000056 |
ribosomal small subunit export from nucleus(GO:0000056) |
| 0.2 |
1.1 |
GO:0006189 |
'de novo' IMP biosynthetic process(GO:0006189) |
| 0.2 |
0.9 |
GO:0006438 |
valyl-tRNA aminoacylation(GO:0006438) |
| 0.2 |
2.2 |
GO:0046051 |
UTP metabolic process(GO:0046051) |
| 0.2 |
0.4 |
GO:0006362 |
transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.2 |
0.7 |
GO:0043988 |
histone H3-S28 phosphorylation(GO:0043988) |
| 0.2 |
0.7 |
GO:1904685 |
positive regulation of metalloendopeptidase activity(GO:1904685) |
| 0.2 |
0.7 |
GO:0043323 |
positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.2 |
2.1 |
GO:0034551 |
respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.2 |
0.6 |
GO:0002940 |
tRNA N2-guanine methylation(GO:0002940) |
| 0.2 |
0.9 |
GO:0006450 |
regulation of translational fidelity(GO:0006450) |
| 0.2 |
1.1 |
GO:0050747 |
positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.2 |
1.5 |
GO:0070458 |
detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.2 |
1.0 |
GO:2000680 |
regulation of rubidium ion transport(GO:2000680) |
| 0.2 |
0.6 |
GO:0071873 |
response to norepinephrine(GO:0071873) |
| 0.2 |
0.6 |
GO:0006285 |
base-excision repair, AP site formation(GO:0006285) |
| 0.2 |
0.6 |
GO:0003100 |
regulation of systemic arterial blood pressure by endothelin(GO:0003100) |
| 0.2 |
1.0 |
GO:0014022 |
neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.2 |
0.8 |
GO:0030576 |
Cajal body organization(GO:0030576) |
| 0.2 |
0.2 |
GO:0046036 |
CTP biosynthetic process(GO:0006241) pyrimidine ribonucleoside triphosphate biosynthetic process(GO:0009209) CTP metabolic process(GO:0046036) |
| 0.2 |
0.2 |
GO:0045084 |
positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.2 |
0.8 |
GO:0019249 |
lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) |
| 0.2 |
0.6 |
GO:0016056 |
rhodopsin mediated signaling pathway(GO:0016056) |
| 0.2 |
0.4 |
GO:0046084 |
adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.2 |
0.6 |
GO:0032430 |
positive regulation of phospholipase A2 activity(GO:0032430) activation of meiosis involved in egg activation(GO:0060466) |
| 0.2 |
1.3 |
GO:0009162 |
deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.2 |
0.8 |
GO:0019509 |
L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.2 |
1.8 |
GO:0010572 |
positive regulation of platelet activation(GO:0010572) |
| 0.2 |
0.6 |
GO:0031394 |
positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.2 |
0.5 |
GO:0021759 |
globus pallidus development(GO:0021759) |
| 0.2 |
0.5 |
GO:0071544 |
diphosphoinositol polyphosphate metabolic process(GO:0071543) diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.2 |
1.4 |
GO:0071907 |
determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.2 |
1.8 |
GO:0051901 |
positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.2 |
0.5 |
GO:0015889 |
cobalamin transport(GO:0015889) |
| 0.2 |
0.7 |
GO:0035624 |
receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.2 |
1.2 |
GO:0060666 |
dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.2 |
0.5 |
GO:0032696 |
negative regulation of interleukin-13 production(GO:0032696) |
| 0.2 |
0.5 |
GO:1904706 |
response to cisplatin(GO:0072718) negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.2 |
1.5 |
GO:0045723 |
positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.2 |
0.5 |
GO:0009193 |
pyrimidine nucleotide catabolic process(GO:0006244) pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.2 |
0.7 |
GO:0071930 |
negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.2 |
0.3 |
GO:2000020 |
positive regulation of male gonad development(GO:2000020) |
| 0.2 |
0.7 |
GO:0042360 |
vitamin E metabolic process(GO:0042360) |
| 0.2 |
0.5 |
GO:0010899 |
regulation of phosphatidylcholine catabolic process(GO:0010899) |
| 0.2 |
0.6 |
GO:0032901 |
positive regulation of neurotrophin production(GO:0032901) |
| 0.2 |
0.3 |
GO:0090286 |
cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.2 |
0.6 |
GO:0070460 |
thyroid-stimulating hormone secretion(GO:0070460) |
| 0.2 |
0.2 |
GO:0060528 |
secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.2 |
0.8 |
GO:0048050 |
post-embryonic eye morphogenesis(GO:0048050) |
| 0.1 |
0.6 |
GO:0018158 |
protein oxidation(GO:0018158) |
| 0.1 |
0.6 |
GO:0032439 |
endosome localization(GO:0032439) |
| 0.1 |
0.4 |
GO:0042421 |
norepinephrine biosynthetic process(GO:0042421) |
| 0.1 |
0.4 |
GO:0021893 |
cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 0.1 |
0.6 |
GO:0060155 |
platelet dense granule organization(GO:0060155) |
| 0.1 |
0.4 |
GO:0072356 |
chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.1 |
0.1 |
GO:0034124 |
regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.1 |
1.0 |
GO:0008616 |
queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.1 |
0.6 |
GO:0017183 |
peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 |
2.7 |
GO:0038092 |
nodal signaling pathway(GO:0038092) |
| 0.1 |
1.7 |
GO:0000463 |
maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 |
0.3 |
GO:2000157 |
regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.1 |
1.4 |
GO:0051103 |
DNA ligation involved in DNA repair(GO:0051103) |
| 0.1 |
0.4 |
GO:0046881 |
positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.1 |
1.0 |
GO:0010273 |
detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.1 |
2.1 |
GO:0045792 |
negative regulation of cell size(GO:0045792) |
| 0.1 |
0.4 |
GO:0051182 |
coenzyme transport(GO:0051182) |
| 0.1 |
0.7 |
GO:0009052 |
pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.1 |
1.2 |
GO:0045198 |
establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 |
0.1 |
GO:0071895 |
odontoblast differentiation(GO:0071895) |
| 0.1 |
0.9 |
GO:0061484 |
hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 |
0.4 |
GO:0042822 |
vitamin B6 metabolic process(GO:0042816) pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.1 |
0.5 |
GO:0035356 |
cellular triglyceride homeostasis(GO:0035356) |
| 0.1 |
0.4 |
GO:0071038 |
U1 snRNA 3'-end processing(GO:0034473) U4 snRNA 3'-end processing(GO:0034475) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent tRNA catabolic process(GO:0071038) |
| 0.1 |
3.4 |
GO:2000060 |
positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.1 |
0.6 |
GO:0002922 |
positive regulation of humoral immune response(GO:0002922) |
| 0.1 |
0.4 |
GO:0042908 |
xenobiotic transport(GO:0042908) |
| 0.1 |
0.1 |
GO:0009153 |
purine deoxyribonucleotide biosynthetic process(GO:0009153) |
| 0.1 |
0.6 |
GO:0040031 |
snRNA modification(GO:0040031) |
| 0.1 |
0.5 |
GO:0051151 |
negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 |
0.4 |
GO:1902219 |
regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.1 |
0.2 |
GO:1902224 |
cellular ketone body metabolic process(GO:0046950) ketone body metabolic process(GO:1902224) |
| 0.1 |
0.2 |
GO:0010643 |
cell communication by chemical coupling(GO:0010643) |
| 0.1 |
0.6 |
GO:0002315 |
marginal zone B cell differentiation(GO:0002315) |
| 0.1 |
0.4 |
GO:0006592 |
ornithine biosynthetic process(GO:0006592) |
| 0.1 |
0.7 |
GO:0043415 |
positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.1 |
0.4 |
GO:0034310 |
primary alcohol catabolic process(GO:0034310) |
| 0.1 |
0.4 |
GO:0010845 |
positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.1 |
0.7 |
GO:0003383 |
apical constriction(GO:0003383) |
| 0.1 |
0.8 |
GO:0061158 |
3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 |
0.3 |
GO:0046032 |
ADP catabolic process(GO:0046032) |
| 0.1 |
1.6 |
GO:0034308 |
primary alcohol metabolic process(GO:0034308) |
| 0.1 |
0.7 |
GO:0051013 |
microtubule severing(GO:0051013) |
| 0.1 |
4.8 |
GO:0006414 |
translational elongation(GO:0006414) |
| 0.1 |
1.4 |
GO:0000244 |
spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 |
0.2 |
GO:0006207 |
'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.1 |
0.3 |
GO:0060729 |
intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 |
0.9 |
GO:0015791 |
polyol transport(GO:0015791) |
| 0.1 |
3.0 |
GO:0071353 |
cellular response to interleukin-4(GO:0071353) |
| 0.1 |
2.9 |
GO:0000027 |
ribosomal large subunit assembly(GO:0000027) |
| 0.1 |
0.4 |
GO:0019355 |
nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 |
0.2 |
GO:0031047 |
gene silencing by RNA(GO:0031047) |
| 0.1 |
0.2 |
GO:0020027 |
hemoglobin metabolic process(GO:0020027) hemoglobin biosynthetic process(GO:0042541) |
| 0.1 |
0.4 |
GO:0070858 |
negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.1 |
0.5 |
GO:0036089 |
cleavage furrow formation(GO:0036089) |
| 0.1 |
0.5 |
GO:1903071 |
positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.1 |
0.4 |
GO:0021586 |
pons maturation(GO:0021586) |
| 0.1 |
0.3 |
GO:1902109 |
negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.1 |
0.3 |
GO:0035973 |
aggrephagy(GO:0035973) |
| 0.1 |
0.3 |
GO:0010757 |
negative regulation of plasminogen activation(GO:0010757) |
| 0.1 |
0.5 |
GO:0090292 |
nuclear migration along microfilament(GO:0031022) nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 |
0.3 |
GO:0045819 |
positive regulation of glycogen catabolic process(GO:0045819) |
| 0.1 |
0.5 |
GO:0006564 |
L-serine biosynthetic process(GO:0006564) |
| 0.1 |
1.3 |
GO:0009143 |
nucleoside triphosphate catabolic process(GO:0009143) |
| 0.1 |
0.4 |
GO:0051725 |
protein de-ADP-ribosylation(GO:0051725) |
| 0.1 |
0.8 |
GO:0033683 |
nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.1 |
0.7 |
GO:0060837 |
blood vessel endothelial cell differentiation(GO:0060837) |
| 0.1 |
0.2 |
GO:0072008 |
regulation of vascular wound healing(GO:0061043) glomerular mesangial cell differentiation(GO:0072008) glomerular mesangial cell development(GO:0072144) |
| 0.1 |
0.3 |
GO:0060437 |
lung growth(GO:0060437) |
| 0.1 |
0.5 |
GO:0032815 |
negative regulation of natural killer cell activation(GO:0032815) |
| 0.1 |
0.6 |
GO:2000672 |
negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 |
0.5 |
GO:0010991 |
regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 |
0.2 |
GO:0030214 |
hyaluronan catabolic process(GO:0030214) |
| 0.1 |
0.4 |
GO:0018343 |
protein farnesylation(GO:0018343) |
| 0.1 |
0.5 |
GO:0015879 |
carnitine transport(GO:0015879) |
| 0.1 |
0.3 |
GO:0043181 |
vacuolar sequestering(GO:0043181) |
| 0.1 |
0.3 |
GO:2000983 |
regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.1 |
0.5 |
GO:0014062 |
regulation of serotonin secretion(GO:0014062) |
| 0.1 |
1.5 |
GO:0006220 |
pyrimidine nucleotide metabolic process(GO:0006220) |
| 0.1 |
0.6 |
GO:0090312 |
positive regulation of protein deacetylation(GO:0090312) |
| 0.1 |
1.3 |
GO:1990403 |
embryonic brain development(GO:1990403) |
| 0.1 |
0.2 |
GO:0001922 |
B-1 B cell homeostasis(GO:0001922) |
| 0.1 |
0.5 |
GO:0006048 |
UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.1 |
1.3 |
GO:0010388 |
protein deneddylation(GO:0000338) cullin deneddylation(GO:0010388) |
| 0.1 |
0.3 |
GO:0046133 |
pyrimidine ribonucleoside catabolic process(GO:0046133) |
| 0.1 |
0.5 |
GO:0034080 |
CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 |
0.2 |
GO:0045080 |
positive regulation of chemokine biosynthetic process(GO:0045080) |
| 0.1 |
0.2 |
GO:0042362 |
fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.1 |
0.6 |
GO:1900194 |
negative regulation of oocyte maturation(GO:1900194) |
| 0.1 |
0.4 |
GO:0032227 |
negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.1 |
0.4 |
GO:0071638 |
positive regulation of T-helper 1 cell differentiation(GO:0045627) negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.1 |
0.3 |
GO:0060743 |
estrous cycle(GO:0044849) epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 |
0.3 |
GO:0010756 |
regulation of plasminogen activation(GO:0010755) positive regulation of plasminogen activation(GO:0010756) |
| 0.1 |
0.3 |
GO:0010626 |
negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.1 |
0.2 |
GO:0035793 |
negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) cell migration involved in kidney development(GO:0035787) cell migration involved in metanephros development(GO:0035788) metanephric mesenchymal cell migration(GO:0035789) positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) regulation of metanephric mesenchymal cell migration(GO:2000589) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.1 |
0.3 |
GO:1904139 |
microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.1 |
0.1 |
GO:0046060 |
dATP metabolic process(GO:0046060) |
| 0.1 |
0.5 |
GO:0006046 |
N-acetylglucosamine catabolic process(GO:0006046) |
| 0.1 |
0.5 |
GO:0034393 |
positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.1 |
0.2 |
GO:0006556 |
S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 |
1.2 |
GO:0043011 |
myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 |
0.2 |
GO:0039530 |
MDA-5 signaling pathway(GO:0039530) |
| 0.1 |
1.2 |
GO:0030213 |
hyaluronan biosynthetic process(GO:0030213) |
| 0.1 |
0.2 |
GO:0070682 |
proteasome regulatory particle assembly(GO:0070682) |
| 0.1 |
0.3 |
GO:0043504 |
mitochondrial DNA repair(GO:0043504) |
| 0.1 |
0.3 |
GO:0000491 |
small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.1 |
0.3 |
GO:0019276 |
UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 |
0.1 |
GO:0010165 |
response to X-ray(GO:0010165) |
| 0.1 |
0.2 |
GO:0090200 |
positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 |
0.2 |
GO:0033499 |
galactose catabolic process via UDP-galactose(GO:0033499) glycolytic process from galactose(GO:0061623) |
| 0.1 |
0.6 |
GO:0015862 |
uridine transport(GO:0015862) |
| 0.1 |
0.6 |
GO:0036506 |
maintenance of unfolded protein(GO:0036506) protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.1 |
0.9 |
GO:0043117 |
positive regulation of vascular permeability(GO:0043117) |
| 0.1 |
0.1 |
GO:0001844 |
protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:0001844) |
| 0.1 |
0.5 |
GO:0046502 |
uroporphyrinogen III metabolic process(GO:0046502) |
| 0.1 |
0.3 |
GO:0034638 |
phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 |
0.3 |
GO:0030656 |
regulation of vitamin metabolic process(GO:0030656) |
| 0.1 |
0.2 |
GO:0034162 |
toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.1 |
0.2 |
GO:0008612 |
peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.1 |
0.2 |
GO:0035162 |
embryonic hemopoiesis(GO:0035162) |
| 0.1 |
0.5 |
GO:0045842 |
positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.1 |
0.3 |
GO:0021562 |
vestibulocochlear nerve development(GO:0021562) |
| 0.1 |
0.5 |
GO:0002903 |
negative regulation of B cell apoptotic process(GO:0002903) |
| 0.1 |
0.4 |
GO:0043951 |
negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 |
0.1 |
GO:0043496 |
regulation of protein homodimerization activity(GO:0043496) |
| 0.1 |
0.7 |
GO:0006104 |
succinyl-CoA metabolic process(GO:0006104) |
| 0.1 |
1.0 |
GO:0042407 |
cristae formation(GO:0042407) |
| 0.1 |
0.4 |
GO:0000480 |
endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 |
0.5 |
GO:0010908 |
regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) myoblast fate commitment(GO:0048625) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 |
0.8 |
GO:0071803 |
positive regulation of podosome assembly(GO:0071803) |
| 0.1 |
2.6 |
GO:0043489 |
RNA stabilization(GO:0043489) |
| 0.1 |
0.2 |
GO:0070375 |
ERK5 cascade(GO:0070375) |
| 0.1 |
0.3 |
GO:1903416 |
response to glycoside(GO:1903416) |
| 0.1 |
0.2 |
GO:0006348 |
chromatin silencing at telomere(GO:0006348) |
| 0.1 |
1.0 |
GO:0042182 |
ketone catabolic process(GO:0042182) |
| 0.1 |
0.6 |
GO:0007440 |
foregut morphogenesis(GO:0007440) embryonic foregut morphogenesis(GO:0048617) |
| 0.1 |
0.2 |
GO:0070625 |
zymogen granule exocytosis(GO:0070625) |
| 0.1 |
0.3 |
GO:0015786 |
UDP-glucose transport(GO:0015786) |
| 0.1 |
0.4 |
GO:0010668 |
ectodermal cell differentiation(GO:0010668) |
| 0.1 |
0.1 |
GO:0071442 |
positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.1 |
0.2 |
GO:0002014 |
vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.1 |
0.8 |
GO:0080111 |
DNA demethylation(GO:0080111) |
| 0.1 |
0.4 |
GO:0090502 |
RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.1 |
1.0 |
GO:0030150 |
protein import into mitochondrial matrix(GO:0030150) |
| 0.1 |
0.3 |
GO:0009227 |
UDP-N-acetylglucosamine catabolic process(GO:0006049) nucleotide-sugar catabolic process(GO:0009227) |
| 0.1 |
0.2 |
GO:0010986 |
positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.1 |
0.7 |
GO:0001516 |
prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.1 |
0.2 |
GO:0019074 |
viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 |
0.7 |
GO:0043206 |
extracellular fibril organization(GO:0043206) |
| 0.1 |
0.1 |
GO:0044332 |
Wnt signaling pathway involved in dorsal/ventral axis specification(GO:0044332) |
| 0.1 |
0.1 |
GO:0007181 |
transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.1 |
0.6 |
GO:0042304 |
regulation of fatty acid biosynthetic process(GO:0042304) |
| 0.1 |
0.3 |
GO:0016480 |
negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.1 |
0.2 |
GO:0019346 |
homoserine metabolic process(GO:0009092) transsulfuration(GO:0019346) |
| 0.1 |
0.2 |
GO:0035521 |
monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 |
0.2 |
GO:0000237 |
leptotene(GO:0000237) |
| 0.1 |
0.2 |
GO:0036265 |
RNA (guanine-N7)-methylation(GO:0036265) |
| 0.1 |
0.2 |
GO:1904729 |
regulation of intestinal cholesterol absorption(GO:0030300) regulation of intestinal absorption(GO:1904478) regulation of intestinal lipid absorption(GO:1904729) |
| 0.1 |
0.2 |
GO:0060120 |
auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.1 |
0.7 |
GO:0006120 |
mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 |
0.5 |
GO:0045541 |
negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 |
0.9 |
GO:0042744 |
hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 |
0.4 |
GO:0003056 |
regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.1 |
0.3 |
GO:0000727 |
double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 |
0.5 |
GO:0090308 |
regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.1 |
0.3 |
GO:1903772 |
regulation of viral budding via host ESCRT complex(GO:1903772) |
| 0.1 |
0.2 |
GO:1904451 |
regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.1 |
0.7 |
GO:0008535 |
respiratory chain complex IV assembly(GO:0008535) |
| 0.1 |
0.7 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.1 |
0.1 |
GO:0007128 |
meiotic prophase I(GO:0007128) prophase(GO:0051324) meiotic cell cycle phase(GO:0098762) meiosis I cell cycle phase(GO:0098764) |
| 0.1 |
0.2 |
GO:0045342 |
MHC class II biosynthetic process(GO:0045342) regulation of MHC class II biosynthetic process(GO:0045346) negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.1 |
0.1 |
GO:0090193 |
positive regulation of glomerulus development(GO:0090193) |
| 0.1 |
0.2 |
GO:0072180 |
mesenchymal cell proliferation involved in ureteric bud development(GO:0072138) mesonephric duct development(GO:0072177) mesonephric duct morphogenesis(GO:0072180) |
| 0.1 |
0.2 |
GO:0000965 |
mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.1 |
0.2 |
GO:0033152 |
immunoglobulin V(D)J recombination(GO:0033152) |
| 0.1 |
0.3 |
GO:0003419 |
growth plate cartilage chondrocyte proliferation(GO:0003419) regulation of growth plate cartilage chondrocyte proliferation(GO:0003420) |
| 0.1 |
0.1 |
GO:1901970 |
positive regulation of mitotic sister chromatid separation(GO:1901970) |
| 0.1 |
0.2 |
GO:0000379 |
tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 |
0.6 |
GO:0006995 |
cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 |
0.1 |
GO:0070309 |
lens fiber cell morphogenesis(GO:0070309) |
| 0.1 |
0.7 |
GO:0001731 |
formation of translation preinitiation complex(GO:0001731) |
| 0.1 |
0.3 |
GO:0090273 |
regulation of somatostatin secretion(GO:0090273) |
| 0.1 |
0.2 |
GO:0006532 |
aspartate biosynthetic process(GO:0006532) |
| 0.1 |
0.9 |
GO:0033630 |
positive regulation of cell adhesion mediated by integrin(GO:0033630) |
| 0.1 |
0.4 |
GO:0007342 |
fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.1 |
1.6 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 |
0.3 |
GO:0006297 |
nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 |
0.3 |
GO:0046602 |
regulation of mitotic centrosome separation(GO:0046602) |
| 0.1 |
0.2 |
GO:0007262 |
STAT protein import into nucleus(GO:0007262) |
| 0.1 |
0.3 |
GO:2001270 |
regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) |
| 0.1 |
0.4 |
GO:1902035 |
positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.1 |
0.3 |
GO:0046598 |
positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 |
0.4 |
GO:0060510 |
Type II pneumocyte differentiation(GO:0060510) |
| 0.1 |
0.3 |
GO:1903553 |
positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.1 |
0.1 |
GO:0034196 |
acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 |
0.2 |
GO:0090241 |
negative regulation of histone H4 acetylation(GO:0090241) |
| 0.1 |
0.3 |
GO:0071712 |
ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 |
0.1 |
GO:2001137 |
positive regulation of endocytic recycling(GO:2001137) |
| 0.1 |
0.2 |
GO:0030827 |
negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.1 |
0.2 |
GO:0030327 |
prenylated protein catabolic process(GO:0030327) |
| 0.1 |
0.6 |
GO:1990173 |
protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.1 |
0.2 |
GO:0009992 |
cellular water homeostasis(GO:0009992) |
| 0.1 |
0.2 |
GO:2000813 |
actin filament uncapping(GO:0051695) negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.1 |
0.2 |
GO:0006833 |
water transport(GO:0006833) |
| 0.1 |
0.2 |
GO:0043144 |
snoRNA processing(GO:0043144) |
| 0.1 |
0.3 |
GO:0086013 |
membrane repolarization during cardiac muscle cell action potential(GO:0086013) |
| 0.1 |
0.2 |
GO:0018298 |
protein-chromophore linkage(GO:0018298) |
| 0.1 |
0.2 |
GO:0045176 |
apical protein localization(GO:0045176) |
| 0.1 |
0.2 |
GO:0061073 |
ciliary body morphogenesis(GO:0061073) |
| 0.1 |
0.5 |
GO:0042989 |
sequestering of actin monomers(GO:0042989) |
| 0.1 |
0.6 |
GO:0000028 |
ribosomal small subunit assembly(GO:0000028) |
| 0.1 |
0.2 |
GO:0070589 |
cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) cell wall biogenesis(GO:0042546) cell wall macromolecule metabolic process(GO:0044036) cell wall macromolecule biosynthetic process(GO:0044038) chain elongation of O-linked mannose residue(GO:0044845) cellular component macromolecule biosynthetic process(GO:0070589) cell wall organization or biogenesis(GO:0071554) |
| 0.1 |
1.4 |
GO:0009142 |
nucleoside triphosphate biosynthetic process(GO:0009142) |
| 0.1 |
0.4 |
GO:0090244 |
Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.0 |
0.2 |
GO:0090073 |
positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 |
0.1 |
GO:0032877 |
positive regulation of DNA endoreduplication(GO:0032877) |
| 0.0 |
0.2 |
GO:0007253 |
cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 |
0.3 |
GO:0032471 |
negative regulation of endoplasmic reticulum calcium ion concentration(GO:0032471) |
| 0.0 |
0.3 |
GO:0032534 |
regulation of microvillus assembly(GO:0032534) |
| 0.0 |
0.2 |
GO:0070459 |
prolactin secretion(GO:0070459) |
| 0.0 |
0.1 |
GO:0060662 |
oviduct development(GO:0060066) tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 |
0.1 |
GO:0035582 |
sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.0 |
0.9 |
GO:0042773 |
ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 |
0.2 |
GO:0050812 |
regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 |
0.7 |
GO:0009083 |
branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 |
0.2 |
GO:0006689 |
ganglioside catabolic process(GO:0006689) |
| 0.0 |
0.1 |
GO:0006097 |
glyoxylate cycle(GO:0006097) |
| 0.0 |
0.1 |
GO:0070889 |
platelet alpha granule organization(GO:0070889) |
| 0.0 |
0.1 |
GO:0000320 |
re-entry into mitotic cell cycle(GO:0000320) |
| 0.0 |
1.3 |
GO:0070979 |
protein K11-linked ubiquitination(GO:0070979) |
| 0.0 |
0.2 |
GO:1901724 |
positive regulation of cell proliferation involved in kidney development(GO:1901724) |
| 0.0 |
0.1 |
GO:0046900 |
tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 |
0.0 |
GO:0010040 |
response to iron(II) ion(GO:0010040) |
| 0.0 |
0.4 |
GO:1902902 |
negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 |
1.5 |
GO:0007218 |
neuropeptide signaling pathway(GO:0007218) |
| 0.0 |
1.0 |
GO:0016578 |
histone deubiquitination(GO:0016578) |
| 0.0 |
0.3 |
GO:2000766 |
negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 |
0.9 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 |
0.7 |
GO:0015812 |
gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 |
0.3 |
GO:0035948 |
positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.0 |
1.0 |
GO:0032008 |
positive regulation of TOR signaling(GO:0032008) |
| 0.0 |
0.2 |
GO:2000628 |
regulation of miRNA metabolic process(GO:2000628) |
| 0.0 |
0.1 |
GO:0098528 |
skeletal muscle fiber differentiation(GO:0098528) |
| 0.0 |
0.3 |
GO:1901857 |
positive regulation of cellular respiration(GO:1901857) |
| 0.0 |
0.2 |
GO:0048069 |
eye pigmentation(GO:0048069) |
| 0.0 |
0.1 |
GO:0070257 |
positive regulation of mucus secretion(GO:0070257) |
| 0.0 |
0.5 |
GO:0043153 |
entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 |
0.2 |
GO:1901843 |
positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.0 |
0.1 |
GO:0015960 |
diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.0 |
0.4 |
GO:0006924 |
activation-induced cell death of T cells(GO:0006924) |
| 0.0 |
0.2 |
GO:0051697 |
protein delipidation(GO:0051697) |
| 0.0 |
0.2 |
GO:0050862 |
positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 |
0.1 |
GO:0007161 |
calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.0 |
0.2 |
GO:0034587 |
piRNA metabolic process(GO:0034587) |
| 0.0 |
0.1 |
GO:0060242 |
contact inhibition(GO:0060242) |
| 0.0 |
0.2 |
GO:0019236 |
response to pheromone(GO:0019236) |
| 0.0 |
1.3 |
GO:0045071 |
negative regulation of viral genome replication(GO:0045071) |
| 0.0 |
0.7 |
GO:0070884 |
regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 |
0.1 |
GO:1901386 |
negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.0 |
0.0 |
GO:0042637 |
catagen(GO:0042637) |
| 0.0 |
0.7 |
GO:2000757 |
negative regulation of peptidyl-lysine acetylation(GO:2000757) |
| 0.0 |
0.4 |
GO:0002021 |
response to dietary excess(GO:0002021) |
| 0.0 |
0.8 |
GO:0042474 |
middle ear morphogenesis(GO:0042474) |
| 0.0 |
0.2 |
GO:0044026 |
DNA hypermethylation(GO:0044026) hypermethylation of CpG island(GO:0044027) |
| 0.0 |
0.4 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 |
0.3 |
GO:0014816 |
skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 |
1.0 |
GO:0018345 |
protein palmitoylation(GO:0018345) |
| 0.0 |
0.2 |
GO:0051365 |
cellular response to potassium ion starvation(GO:0051365) |
| 0.0 |
0.8 |
GO:0060445 |
branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 |
0.8 |
GO:0055012 |
ventricular cardiac muscle cell differentiation(GO:0055012) |
| 0.0 |
0.3 |
GO:0010884 |
positive regulation of lipid storage(GO:0010884) |
| 0.0 |
0.3 |
GO:0042760 |
very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 |
0.2 |
GO:0008054 |
negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) |
| 0.0 |
0.1 |
GO:0097211 |
response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 |
0.5 |
GO:0051451 |
myoblast migration(GO:0051451) |
| 0.0 |
0.2 |
GO:0006668 |
sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.0 |
0.4 |
GO:1902018 |
negative regulation of cilium assembly(GO:1902018) |
| 0.0 |
0.1 |
GO:0060192 |
negative regulation of lipase activity(GO:0060192) |
| 0.0 |
0.1 |
GO:0019348 |
polyprenol metabolic process(GO:0016093) dolichol metabolic process(GO:0019348) |
| 0.0 |
0.3 |
GO:0034720 |
histone H3-K4 demethylation(GO:0034720) |
| 0.0 |
0.1 |
GO:0000957 |
mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 |
0.3 |
GO:0030490 |
maturation of SSU-rRNA(GO:0030490) |
| 0.0 |
0.1 |
GO:0019695 |
choline metabolic process(GO:0019695) |
| 0.0 |
0.1 |
GO:0015705 |
iodide transport(GO:0015705) |
| 0.0 |
0.9 |
GO:2000279 |
negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.0 |
0.0 |
GO:0010871 |
negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.0 |
0.1 |
GO:0070327 |
thyroid hormone transport(GO:0070327) |
| 0.0 |
0.3 |
GO:0071451 |
cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) |
| 0.0 |
1.9 |
GO:0006446 |
regulation of translational initiation(GO:0006446) |
| 0.0 |
0.1 |
GO:0006116 |
NADH oxidation(GO:0006116) |
| 0.0 |
0.4 |
GO:0021889 |
olfactory bulb interneuron differentiation(GO:0021889) |
| 0.0 |
0.1 |
GO:0090343 |
positive regulation of cell aging(GO:0090343) positive regulation of cellular senescence(GO:2000774) |
| 0.0 |
0.8 |
GO:0033235 |
positive regulation of protein sumoylation(GO:0033235) |
| 0.0 |
0.1 |
GO:0003245 |
cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.0 |
0.2 |
GO:0018401 |
peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 |
0.2 |
GO:1900028 |
negative regulation of ruffle assembly(GO:1900028) |
| 0.0 |
0.9 |
GO:0042273 |
ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 |
0.4 |
GO:0071229 |
cellular response to acid chemical(GO:0071229) |
| 0.0 |
0.2 |
GO:0034227 |
tRNA thio-modification(GO:0034227) |
| 0.0 |
0.4 |
GO:0009072 |
aromatic amino acid family metabolic process(GO:0009072) |
| 0.0 |
0.1 |
GO:0035701 |
hematopoietic stem cell migration(GO:0035701) |
| 0.0 |
0.1 |
GO:1904529 |
regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.0 |
0.6 |
GO:0016558 |
protein import into peroxisome matrix(GO:0016558) |
| 0.0 |
0.1 |
GO:0042732 |
D-xylose metabolic process(GO:0042732) |
| 0.0 |
0.1 |
GO:0090071 |
negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 |
0.1 |
GO:0033089 |
positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 |
0.1 |
GO:0046826 |
negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 |
0.1 |
GO:0050774 |
negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 |
0.4 |
GO:0003416 |
endochondral bone growth(GO:0003416) |
| 0.0 |
0.4 |
GO:0043552 |
positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 |
0.2 |
GO:0070317 |
negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 |
0.1 |
GO:0032423 |
regulation of mismatch repair(GO:0032423) |
| 0.0 |
0.5 |
GO:0045672 |
positive regulation of osteoclast differentiation(GO:0045672) |
| 0.0 |
0.1 |
GO:0031536 |
positive regulation of exit from mitosis(GO:0031536) |
| 0.0 |
0.1 |
GO:0010835 |
regulation of protein ADP-ribosylation(GO:0010835) |
| 0.0 |
0.5 |
GO:0031115 |
negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 |
0.1 |
GO:0009298 |
GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 |
0.1 |
GO:0042748 |
circadian sleep/wake cycle, non-REM sleep(GO:0042748) |
| 0.0 |
0.4 |
GO:0060707 |
trophoblast giant cell differentiation(GO:0060707) |
| 0.0 |
0.2 |
GO:0036035 |
osteoclast development(GO:0036035) |
| 0.0 |
0.2 |
GO:0072643 |
interferon-gamma secretion(GO:0072643) |
| 0.0 |
0.0 |
GO:1904008 |
cellular response to salt(GO:1902075) response to monosodium glutamate(GO:1904008) cellular response to monosodium glutamate(GO:1904009) |
| 0.0 |
0.8 |
GO:0000462 |
maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 |
0.3 |
GO:0031167 |
rRNA methylation(GO:0031167) |
| 0.0 |
0.4 |
GO:0050850 |
positive regulation of calcium-mediated signaling(GO:0050850) |
| 0.0 |
0.1 |
GO:0072148 |
epithelial cell fate commitment(GO:0072148) |
| 0.0 |
0.1 |
GO:2000465 |
regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.0 |
0.1 |
GO:1901301 |
regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 |
0.3 |
GO:0061179 |
negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 |
0.2 |
GO:0034724 |
DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 |
0.0 |
GO:0032753 |
positive regulation of interleukin-4 production(GO:0032753) |
| 0.0 |
0.6 |
GO:0031648 |
protein destabilization(GO:0031648) |
| 0.0 |
0.1 |
GO:0000430 |
regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 |
0.3 |
GO:0014883 |
transition between fast and slow fiber(GO:0014883) |
| 0.0 |
0.8 |
GO:0045214 |
sarcomere organization(GO:0045214) |
| 0.0 |
0.1 |
GO:1901165 |
positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 |
0.3 |
GO:0000290 |
deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 |
0.4 |
GO:0035269 |
protein O-linked mannosylation(GO:0035269) |
| 0.0 |
0.2 |
GO:0061051 |
positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.0 |
0.1 |
GO:2000662 |
interleukin-5 secretion(GO:0072603) interleukin-13 secretion(GO:0072611) regulation of interleukin-5 secretion(GO:2000662) positive regulation of interleukin-5 secretion(GO:2000664) regulation of interleukin-13 secretion(GO:2000665) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.0 |
0.2 |
GO:1902231 |
positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 |
0.1 |
GO:0045054 |
constitutive secretory pathway(GO:0045054) |
| 0.0 |
0.0 |
GO:2000642 |
negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.0 |
0.1 |
GO:1903361 |
protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 |
0.1 |
GO:0031339 |
negative regulation of vesicle fusion(GO:0031339) |
| 0.0 |
0.4 |
GO:2000179 |
positive regulation of neural precursor cell proliferation(GO:2000179) |
| 0.0 |
0.1 |
GO:0032020 |
ISG15-protein conjugation(GO:0032020) |
| 0.0 |
0.2 |
GO:0030578 |
PML body organization(GO:0030578) |
| 0.0 |
0.2 |
GO:0044872 |
lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 |
0.7 |
GO:0045604 |
regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 |
0.6 |
GO:0008584 |
male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.0 |
0.2 |
GO:0050901 |
leukocyte tethering or rolling(GO:0050901) |
| 0.0 |
0.1 |
GO:2000310 |
regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 |
0.1 |
GO:2000819 |
regulation of nucleotide-excision repair(GO:2000819) |
| 0.0 |
0.1 |
GO:0009068 |
aspartate family amino acid catabolic process(GO:0009068) |
| 0.0 |
0.2 |
GO:0070301 |
cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 |
0.3 |
GO:0010614 |
negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 |
0.2 |
GO:0031468 |
nuclear envelope reassembly(GO:0031468) |
| 0.0 |
0.4 |
GO:0045943 |
positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 |
0.2 |
GO:0033603 |
positive regulation of dopamine secretion(GO:0033603) |
| 0.0 |
0.6 |
GO:0016575 |
histone deacetylation(GO:0016575) |
| 0.0 |
0.0 |
GO:0014046 |
dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 |
0.2 |
GO:2001238 |
positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 |
0.1 |
GO:0034143 |
regulation of toll-like receptor 4 signaling pathway(GO:0034143) |
| 0.0 |
0.2 |
GO:0071157 |
negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 |
0.3 |
GO:0042104 |
positive regulation of activated T cell proliferation(GO:0042104) |
| 0.0 |
0.9 |
GO:0018279 |
protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 |
0.0 |
GO:0019388 |
galactose catabolic process(GO:0019388) |
| 0.0 |
0.2 |
GO:0006855 |
drug transmembrane transport(GO:0006855) |
| 0.0 |
0.2 |
GO:0019368 |
fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 |
0.2 |
GO:0032525 |
somite rostral/caudal axis specification(GO:0032525) |
| 0.0 |
0.0 |
GO:0009081 |
branched-chain amino acid metabolic process(GO:0009081) |
| 0.0 |
0.0 |
GO:0043379 |
memory T cell differentiation(GO:0043379) |
| 0.0 |
0.2 |
GO:0034498 |
early endosome to Golgi transport(GO:0034498) |
| 0.0 |
0.1 |
GO:0090166 |
Golgi disassembly(GO:0090166) |
| 0.0 |
0.1 |
GO:0048934 |
peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 |
0.4 |
GO:0001967 |
suckling behavior(GO:0001967) |
| 0.0 |
0.1 |
GO:0003062 |
regulation of heart rate by chemical signal(GO:0003062) positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 |
0.3 |
GO:0048147 |
negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 |
0.5 |
GO:0090162 |
establishment of epithelial cell polarity(GO:0090162) |
| 0.0 |
0.4 |
GO:0002181 |
cytoplasmic translation(GO:0002181) |
| 0.0 |
0.2 |
GO:0032793 |
positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 |
0.2 |
GO:1900262 |
regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 |
0.3 |
GO:0031340 |
positive regulation of vesicle fusion(GO:0031340) |
| 0.0 |
0.0 |
GO:0070542 |
response to fatty acid(GO:0070542) |
| 0.0 |
0.0 |
GO:0010759 |
positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 |
0.1 |
GO:0007258 |
JUN phosphorylation(GO:0007258) |
| 0.0 |
0.1 |
GO:1901070 |
guanosine-containing compound biosynthetic process(GO:1901070) |
| 0.0 |
0.3 |
GO:0043248 |
proteasome assembly(GO:0043248) |
| 0.0 |
0.1 |
GO:0015888 |
thiamine transport(GO:0015888) |
| 0.0 |
0.1 |
GO:0021506 |
anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.0 |
0.1 |
GO:0006659 |
phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 |
0.1 |
GO:0051150 |
regulation of smooth muscle cell differentiation(GO:0051150) |
| 0.0 |
0.1 |
GO:0035752 |
lysosomal lumen pH elevation(GO:0035752) |
| 0.0 |
0.1 |
GO:0045741 |
positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 |
0.1 |
GO:0006108 |
malate metabolic process(GO:0006108) |
| 0.0 |
0.2 |
GO:0046085 |
adenosine metabolic process(GO:0046085) |
| 0.0 |
0.0 |
GO:0000972 |
transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 |
0.9 |
GO:0010717 |
regulation of epithelial to mesenchymal transition(GO:0010717) |
| 0.0 |
0.3 |
GO:0042059 |
negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.0 |
0.1 |
GO:0071763 |
nuclear membrane organization(GO:0071763) |
| 0.0 |
0.1 |
GO:0050882 |
voluntary musculoskeletal movement(GO:0050882) |
| 0.0 |
0.2 |
GO:0071392 |
cellular response to estradiol stimulus(GO:0071392) |
| 0.0 |
0.0 |
GO:0045085 |
negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 |
0.1 |
GO:0006527 |
arginine catabolic process(GO:0006527) |
| 0.0 |
0.1 |
GO:0034122 |
negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.0 |
0.8 |
GO:2000649 |
regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 |
0.1 |
GO:0042538 |
hyperosmotic salinity response(GO:0042538) |
| 0.0 |
0.1 |
GO:0071340 |
skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 |
0.0 |
GO:0008355 |
olfactory learning(GO:0008355) |
| 0.0 |
0.1 |
GO:0003254 |
regulation of membrane depolarization(GO:0003254) |
| 0.0 |
0.1 |
GO:0031274 |
regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 |
0.3 |
GO:0032981 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 |
0.1 |
GO:1904715 |
negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.0 |
0.2 |
GO:0044243 |
collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 |
0.1 |
GO:2000253 |
positive regulation of feeding behavior(GO:2000253) |
| 0.0 |
0.1 |
GO:0032780 |
negative regulation of ATPase activity(GO:0032780) |
| 0.0 |
0.0 |
GO:0031584 |
activation of phospholipase D activity(GO:0031584) |
| 0.0 |
0.1 |
GO:0071493 |
cellular response to UV-B(GO:0071493) |
| 0.0 |
0.1 |
GO:0018026 |
peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 |
0.1 |
GO:0051491 |
positive regulation of filopodium assembly(GO:0051491) |
| 0.0 |
0.0 |
GO:0000715 |
nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 |
0.0 |
GO:0007354 |
zygotic determination of anterior/posterior axis, embryo(GO:0007354) |
| 0.0 |
0.1 |
GO:0006741 |
NADP biosynthetic process(GO:0006741) |
| 0.0 |
0.4 |
GO:0046856 |
phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 |
0.3 |
GO:0008637 |
apoptotic mitochondrial changes(GO:0008637) |
| 0.0 |
0.1 |
GO:0030263 |
apoptotic chromosome condensation(GO:0030263) |
| 0.0 |
0.3 |
GO:0001913 |
T cell mediated cytotoxicity(GO:0001913) |
| 0.0 |
1.8 |
GO:0006626 |
protein targeting to mitochondrion(GO:0006626) |
| 0.0 |
0.1 |
GO:0015909 |
long-chain fatty acid transport(GO:0015909) |
| 0.0 |
0.1 |
GO:0043687 |
post-translational protein modification(GO:0043687) |
| 0.0 |
0.0 |
GO:2000254 |
regulation of male germ cell proliferation(GO:2000254) |
| 0.0 |
0.2 |
GO:0071260 |
cellular response to mechanical stimulus(GO:0071260) |
| 0.0 |
0.2 |
GO:0070527 |
platelet aggregation(GO:0070527) |
| 0.0 |
0.2 |
GO:0017144 |
drug metabolic process(GO:0017144) |
| 0.0 |
0.1 |
GO:1904321 |
response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 |
0.1 |
GO:0046339 |
diacylglycerol metabolic process(GO:0046339) |
| 0.0 |
0.1 |
GO:0042790 |
transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.0 |
0.1 |
GO:0001573 |
ganglioside metabolic process(GO:0001573) |
| 0.0 |
0.1 |
GO:0046415 |
urate metabolic process(GO:0046415) |
| 0.0 |
0.2 |
GO:0016486 |
peptide hormone processing(GO:0016486) |
| 0.0 |
1.0 |
GO:0007405 |
neuroblast proliferation(GO:0007405) |
| 0.0 |
0.1 |
GO:0006369 |
termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 |
0.1 |
GO:0045725 |
positive regulation of glycogen biosynthetic process(GO:0045725) positive regulation of glycogen metabolic process(GO:0070875) |
| 0.0 |
0.5 |
GO:0048013 |
ephrin receptor signaling pathway(GO:0048013) |
| 0.0 |
0.1 |
GO:0071156 |
regulation of cell cycle arrest(GO:0071156) |
| 0.0 |
0.6 |
GO:0006891 |
intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 |
0.1 |
GO:0061098 |
positive regulation of protein tyrosine kinase activity(GO:0061098) |
| 0.0 |
0.1 |
GO:0071361 |
cellular response to ethanol(GO:0071361) |
| 0.0 |
0.1 |
GO:0006729 |
tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 |
0.1 |
GO:0033299 |
secretion of lysosomal enzymes(GO:0033299) |
| 0.0 |
0.1 |
GO:0000707 |
meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 |
0.2 |
GO:0071257 |
cellular response to electrical stimulus(GO:0071257) |
| 0.0 |
0.5 |
GO:0032091 |
negative regulation of protein binding(GO:0032091) |
| 0.0 |
0.0 |
GO:0045075 |
interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 |
0.1 |
GO:0098838 |
reduced folate transmembrane transport(GO:0098838) |
| 0.0 |
0.2 |
GO:0050868 |
negative regulation of T cell activation(GO:0050868) |
| 0.0 |
0.2 |
GO:0032402 |
melanosome transport(GO:0032402) |
| 0.0 |
0.1 |
GO:0021796 |
cerebral cortex regionalization(GO:0021796) |
| 0.0 |
0.1 |
GO:2000096 |
positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 |
0.3 |
GO:0006890 |
retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 |
0.0 |
GO:0043383 |
negative T cell selection(GO:0043383) negative thymic T cell selection(GO:0045060) |
| 0.0 |
0.2 |
GO:0033627 |
cell adhesion mediated by integrin(GO:0033627) |
| 0.0 |
0.1 |
GO:0051016 |
barbed-end actin filament capping(GO:0051016) |
| 0.0 |
0.1 |
GO:0043137 |
DNA replication, Okazaki fragment processing(GO:0033567) DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 |
0.2 |
GO:0045787 |
positive regulation of cell cycle(GO:0045787) |
| 0.0 |
0.1 |
GO:0060914 |
heart formation(GO:0060914) |
| 0.0 |
0.2 |
GO:0046677 |
response to antibiotic(GO:0046677) |
| 0.0 |
0.7 |
GO:0042098 |
T cell proliferation(GO:0042098) |
| 0.0 |
0.1 |
GO:0008063 |
Toll signaling pathway(GO:0008063) |
| 0.0 |
0.1 |
GO:0035845 |
photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 |
0.0 |
GO:0015817 |
histidine transport(GO:0015817) |
| 0.0 |
0.2 |
GO:0007140 |
male meiosis(GO:0007140) |
| 0.0 |
0.1 |
GO:0090316 |
positive regulation of intracellular protein transport(GO:0090316) |
| 0.0 |
0.1 |
GO:0006003 |
fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 |
0.1 |
GO:0006636 |
unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.0 |
0.0 |
GO:0046015 |
regulation of transcription by glucose(GO:0046015) |
| 0.0 |
0.2 |
GO:0000038 |
very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 |
0.0 |
GO:2000210 |
positive regulation of anoikis(GO:2000210) |
| 0.0 |
0.2 |
GO:0030261 |
chromosome condensation(GO:0030261) |
| 0.0 |
0.3 |
GO:0007200 |
phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 |
0.1 |
GO:0060441 |
epithelial tube branching involved in lung morphogenesis(GO:0060441) |
| 0.0 |
0.1 |
GO:0055008 |
cardiac muscle tissue morphogenesis(GO:0055008) muscle tissue morphogenesis(GO:0060415) |
| 0.0 |
0.1 |
GO:0006398 |
mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 |
0.5 |
GO:0006413 |
translational initiation(GO:0006413) |
| 0.0 |
0.3 |
GO:0030901 |
midbrain development(GO:0030901) |
| 0.0 |
0.3 |
GO:0034723 |
DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 |
0.1 |
GO:0016925 |
protein sumoylation(GO:0016925) |
| 0.0 |
0.1 |
GO:0051131 |
chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 |
0.0 |
GO:0042149 |
cellular response to glucose starvation(GO:0042149) |
| 0.0 |
0.1 |
GO:0060632 |
regulation of microtubule-based movement(GO:0060632) |
| 0.0 |
0.1 |
GO:0009303 |
rRNA transcription(GO:0009303) |
| 0.0 |
0.4 |
GO:0043966 |
histone H3 acetylation(GO:0043966) |
| 0.0 |
0.0 |
GO:0039536 |
negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 |
0.1 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 |
0.1 |
GO:0051683 |
establishment of Golgi localization(GO:0051683) |
| 0.0 |
0.0 |
GO:0060715 |
syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.0 |
0.1 |
GO:0032486 |
Rap protein signal transduction(GO:0032486) |