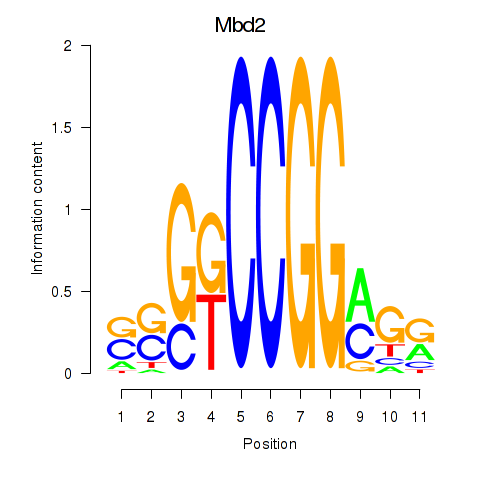
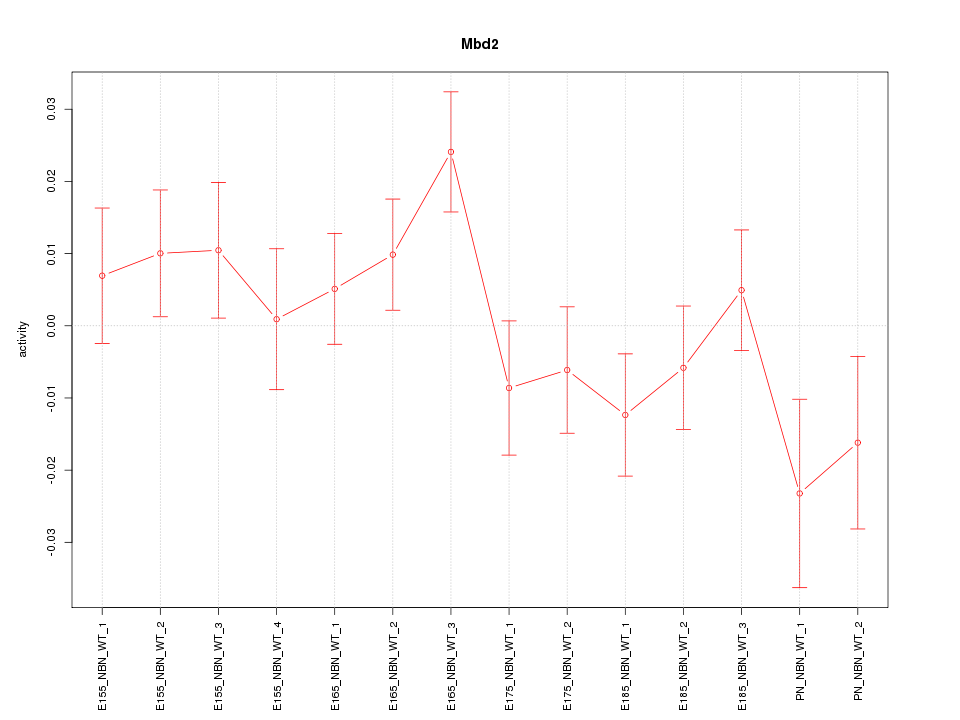
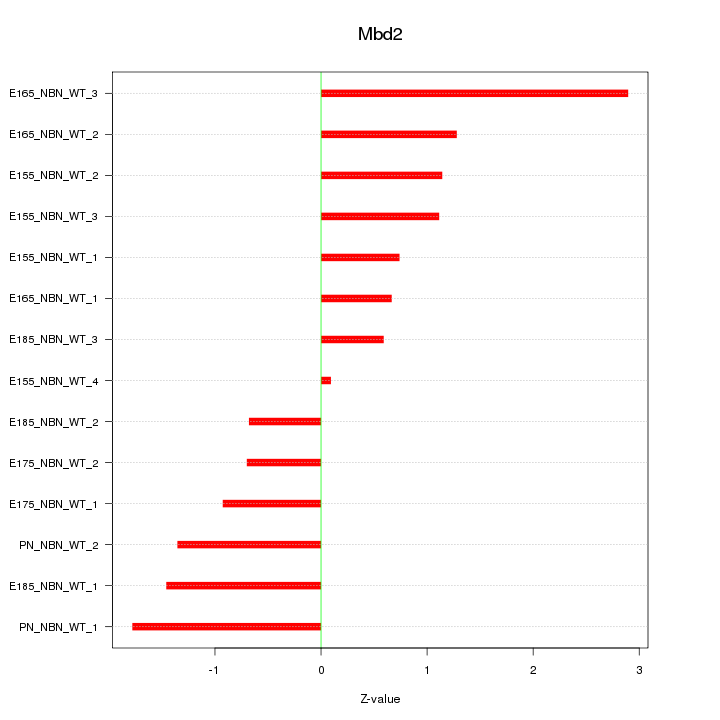
| 1.8 |
11.1 |
GO:0021773 |
striatal medium spiny neuron differentiation(GO:0021773) |
| 1.5 |
4.4 |
GO:1900133 |
renin secretion into blood stream(GO:0002001) negative regulation of urine volume(GO:0035811) regulation of renin secretion into blood stream(GO:1900133) |
| 1.0 |
3.1 |
GO:0035604 |
fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) coronal suture morphogenesis(GO:0060365) squamous basal epithelial stem cell differentiation involved in prostate gland acinus development(GO:0060529) |
| 0.7 |
3.6 |
GO:0036324 |
vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.7 |
2.0 |
GO:0072070 |
establishment or maintenance of polarity of embryonic epithelium(GO:0016332) loop of Henle development(GO:0072070) metanephric loop of Henle development(GO:0072236) |
| 0.6 |
1.9 |
GO:0035574 |
histone H4-K20 demethylation(GO:0035574) |
| 0.5 |
1.5 |
GO:0045110 |
intermediate filament bundle assembly(GO:0045110) |
| 0.5 |
1.9 |
GO:0014846 |
esophagus smooth muscle contraction(GO:0014846) |
| 0.4 |
1.6 |
GO:0007228 |
positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.4 |
1.5 |
GO:0032227 |
negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.4 |
0.7 |
GO:0060743 |
epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.3 |
1.0 |
GO:0060023 |
soft palate development(GO:0060023) |
| 0.3 |
1.0 |
GO:0032877 |
positive regulation of DNA endoreduplication(GO:0032877) |
| 0.3 |
1.0 |
GO:0018103 |
protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.3 |
0.8 |
GO:0044849 |
estrous cycle(GO:0044849) |
| 0.3 |
0.8 |
GO:2000744 |
anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.3 |
1.1 |
GO:0045896 |
regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.3 |
2.3 |
GO:0021830 |
interneuron migration from the subpallium to the cortex(GO:0021830) |
| 0.2 |
0.7 |
GO:0015772 |
disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.2 |
0.7 |
GO:0086046 |
membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.2 |
0.7 |
GO:0019401 |
alditol biosynthetic process(GO:0019401) |
| 0.2 |
1.0 |
GO:0016266 |
O-glycan processing(GO:0016266) |
| 0.2 |
0.6 |
GO:1901552 |
positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.2 |
0.6 |
GO:0021849 |
neuroblast division in subventricular zone(GO:0021849) |
| 0.2 |
0.8 |
GO:1903378 |
positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.2 |
1.4 |
GO:0038031 |
non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.2 |
3.5 |
GO:0050650 |
chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.1 |
1.6 |
GO:1901250 |
regulation of lung goblet cell differentiation(GO:1901249) negative regulation of lung goblet cell differentiation(GO:1901250) |
| 0.1 |
0.7 |
GO:0090666 |
scaRNA localization to Cajal body(GO:0090666) |
| 0.1 |
0.7 |
GO:0071896 |
protein localization to adherens junction(GO:0071896) |
| 0.1 |
0.5 |
GO:2001137 |
positive regulation of endocytic recycling(GO:2001137) |
| 0.1 |
0.6 |
GO:0070125 |
mitochondrial translational elongation(GO:0070125) |
| 0.1 |
0.8 |
GO:0001887 |
selenium compound metabolic process(GO:0001887) |
| 0.1 |
0.4 |
GO:1902174 |
positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.1 |
0.3 |
GO:1901097 |
negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 |
0.3 |
GO:0072554 |
arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) endothelial cell fate specification(GO:0060847) Notch signaling pathway involved in arterial endothelial cell fate commitment(GO:0060853) blood vessel lumenization(GO:0072554) blood vessel endothelial cell fate specification(GO:0097101) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of heart induction(GO:1901321) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.1 |
0.4 |
GO:0043973 |
histone H3-K4 acetylation(GO:0043973) |
| 0.1 |
0.6 |
GO:0070417 |
cellular response to cold(GO:0070417) |
| 0.1 |
0.5 |
GO:0070365 |
hepatocyte differentiation(GO:0070365) |
| 0.1 |
1.4 |
GO:0007190 |
activation of adenylate cyclase activity(GO:0007190) |
| 0.1 |
2.4 |
GO:2000311 |
regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 |
0.5 |
GO:0071955 |
recycling endosome to Golgi transport(GO:0071955) |
| 0.1 |
1.0 |
GO:0090435 |
protein localization to nuclear envelope(GO:0090435) |
| 0.1 |
0.5 |
GO:0035087 |
siRNA loading onto RISC involved in RNA interference(GO:0035087) |
| 0.1 |
0.4 |
GO:0070535 |
histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.1 |
1.2 |
GO:0032486 |
Rap protein signal transduction(GO:0032486) |
| 0.1 |
2.2 |
GO:0007020 |
microtubule nucleation(GO:0007020) |
| 0.1 |
0.3 |
GO:0034184 |
positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.1 |
1.8 |
GO:0006607 |
NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 |
0.8 |
GO:0030322 |
stabilization of membrane potential(GO:0030322) |
| 0.1 |
0.3 |
GO:0007352 |
zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 |
1.2 |
GO:1900454 |
positive regulation of long term synaptic depression(GO:1900454) |
| 0.1 |
0.5 |
GO:0016480 |
negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.1 |
0.6 |
GO:0000963 |
mitochondrial RNA processing(GO:0000963) |
| 0.1 |
0.2 |
GO:0010626 |
negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.1 |
0.3 |
GO:0048069 |
eye pigmentation(GO:0048069) |
| 0.1 |
0.3 |
GO:0046898 |
response to cycloheximide(GO:0046898) |
| 0.1 |
0.3 |
GO:0072675 |
osteoclast fusion(GO:0072675) |
| 0.1 |
0.3 |
GO:0070459 |
prolactin secretion(GO:0070459) |
| 0.1 |
0.6 |
GO:0060059 |
embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.1 |
0.2 |
GO:0003162 |
atrioventricular node development(GO:0003162) |
| 0.1 |
0.6 |
GO:0070141 |
response to UV-A(GO:0070141) |
| 0.1 |
0.5 |
GO:0008343 |
adult feeding behavior(GO:0008343) |
| 0.1 |
0.8 |
GO:0051764 |
actin crosslink formation(GO:0051764) |
| 0.1 |
0.3 |
GO:0060178 |
regulation of exocyst localization(GO:0060178) |
| 0.1 |
0.3 |
GO:0035331 |
negative regulation of hippo signaling(GO:0035331) |
| 0.1 |
0.6 |
GO:0034498 |
early endosome to Golgi transport(GO:0034498) |
| 0.1 |
0.7 |
GO:2000480 |
negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 |
0.3 |
GO:0021993 |
initiation of neural tube closure(GO:0021993) |
| 0.1 |
0.2 |
GO:0010499 |
proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 |
0.2 |
GO:0015882 |
L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.1 |
0.2 |
GO:0009446 |
putrescine biosynthetic process(GO:0009446) |
| 0.1 |
1.6 |
GO:0071353 |
cellular response to interleukin-4(GO:0071353) |
| 0.1 |
0.8 |
GO:0045671 |
negative regulation of osteoclast differentiation(GO:0045671) |
| 0.1 |
0.3 |
GO:0070544 |
histone H3-K36 demethylation(GO:0070544) |
| 0.1 |
0.4 |
GO:0007296 |
vitellogenesis(GO:0007296) |
| 0.0 |
1.0 |
GO:0098743 |
cartilage condensation(GO:0001502) cell aggregation(GO:0098743) |
| 0.0 |
0.3 |
GO:0072610 |
interleukin-12 secretion(GO:0072610) regulation of interleukin-12 secretion(GO:2001182) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 |
0.2 |
GO:0050812 |
regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 |
0.1 |
GO:1902953 |
positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.0 |
0.1 |
GO:0002024 |
diet induced thermogenesis(GO:0002024) adaptive thermogenesis(GO:1990845) |
| 0.0 |
0.5 |
GO:0043252 |
prostaglandin transport(GO:0015732) sodium-independent organic anion transport(GO:0043252) |
| 0.0 |
0.4 |
GO:0006370 |
7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 |
0.5 |
GO:0033148 |
positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 |
0.1 |
GO:1904861 |
excitatory synapse assembly(GO:1904861) |
| 0.0 |
0.2 |
GO:0010668 |
ectodermal cell differentiation(GO:0010668) |
| 0.0 |
0.3 |
GO:0046541 |
saliva secretion(GO:0046541) |
| 0.0 |
0.2 |
GO:0090148 |
protein hexamerization(GO:0034214) membrane fission(GO:0090148) |
| 0.0 |
0.8 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 |
0.4 |
GO:0071763 |
nuclear membrane organization(GO:0071763) |
| 0.0 |
0.1 |
GO:0010991 |
regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 |
0.3 |
GO:1902231 |
positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 |
0.5 |
GO:0090141 |
positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 |
0.2 |
GO:0046292 |
formaldehyde metabolic process(GO:0046292) |
| 0.0 |
0.6 |
GO:0031340 |
positive regulation of vesicle fusion(GO:0031340) |
| 0.0 |
1.1 |
GO:0001964 |
startle response(GO:0001964) |
| 0.0 |
0.2 |
GO:0005981 |
regulation of glycogen catabolic process(GO:0005981) |
| 0.0 |
0.2 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 |
0.2 |
GO:0018026 |
peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 |
0.1 |
GO:0071692 |
protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) |
| 0.0 |
0.3 |
GO:0048386 |
positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 |
0.3 |
GO:0051967 |
negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 |
0.4 |
GO:0001516 |
prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 |
0.2 |
GO:0006398 |
mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 |
0.2 |
GO:0033564 |
anterior/posterior axon guidance(GO:0033564) |
| 0.0 |
0.3 |
GO:0033574 |
positive regulation of neurotransmitter secretion(GO:0001956) response to testosterone(GO:0033574) |
| 0.0 |
0.2 |
GO:0061084 |
negative regulation of protein refolding(GO:0061084) |
| 0.0 |
0.4 |
GO:0097094 |
craniofacial suture morphogenesis(GO:0097094) |
| 0.0 |
0.4 |
GO:0090263 |
positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 |
0.1 |
GO:0007217 |
tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 |
0.4 |
GO:0060693 |
regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.0 |
0.4 |
GO:0030512 |
negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 |
0.4 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 |
0.2 |
GO:0045040 |
protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 |
0.1 |
GO:0070562 |
regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 |
0.1 |
GO:2001025 |
positive regulation of response to drug(GO:2001025) |
| 0.0 |
0.3 |
GO:0032957 |
inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 |
2.6 |
GO:0051028 |
mRNA transport(GO:0051028) |
| 0.0 |
0.1 |
GO:0060355 |
positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 |
1.3 |
GO:0046626 |
regulation of insulin receptor signaling pathway(GO:0046626) |
| 0.0 |
0.4 |
GO:0048505 |
regulation of timing of cell differentiation(GO:0048505) |
| 0.0 |
0.3 |
GO:1903337 |
positive regulation of vacuolar transport(GO:1903337) |
| 0.0 |
0.1 |
GO:1901165 |
positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 |
0.1 |
GO:0044027 |
hypermethylation of CpG island(GO:0044027) |
| 0.0 |
0.7 |
GO:0010830 |
regulation of myotube differentiation(GO:0010830) |
| 0.0 |
0.2 |
GO:0009404 |
toxin metabolic process(GO:0009404) |
| 0.0 |
0.3 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 0.0 |
0.0 |
GO:0042699 |
follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 |
0.5 |
GO:1901385 |
regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 |
0.6 |
GO:0035774 |
positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.0 |
0.4 |
GO:0035335 |
peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 |
0.5 |
GO:0061099 |
negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 |
0.4 |
GO:0030866 |
cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 |
0.5 |
GO:0000184 |
nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 |
0.1 |
GO:0019673 |
GDP-mannose metabolic process(GO:0019673) |
| 0.0 |
0.1 |
GO:0042535 |
positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.0 |
0.1 |
GO:0021960 |
anterior commissure morphogenesis(GO:0021960) |
| 0.0 |
0.2 |
GO:0035269 |
protein O-linked mannosylation(GO:0035269) |
| 0.0 |
0.2 |
GO:0001967 |
suckling behavior(GO:0001967) |
| 0.0 |
0.6 |
GO:0035418 |
protein localization to synapse(GO:0035418) |
| 0.0 |
0.3 |
GO:0050982 |
detection of mechanical stimulus(GO:0050982) |
| 0.0 |
0.2 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 |
0.1 |
GO:0033182 |
regulation of histone ubiquitination(GO:0033182) |
| 0.0 |
0.2 |
GO:0043248 |
proteasome assembly(GO:0043248) |