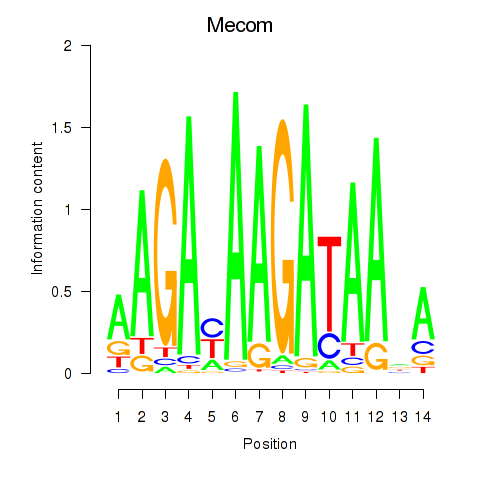
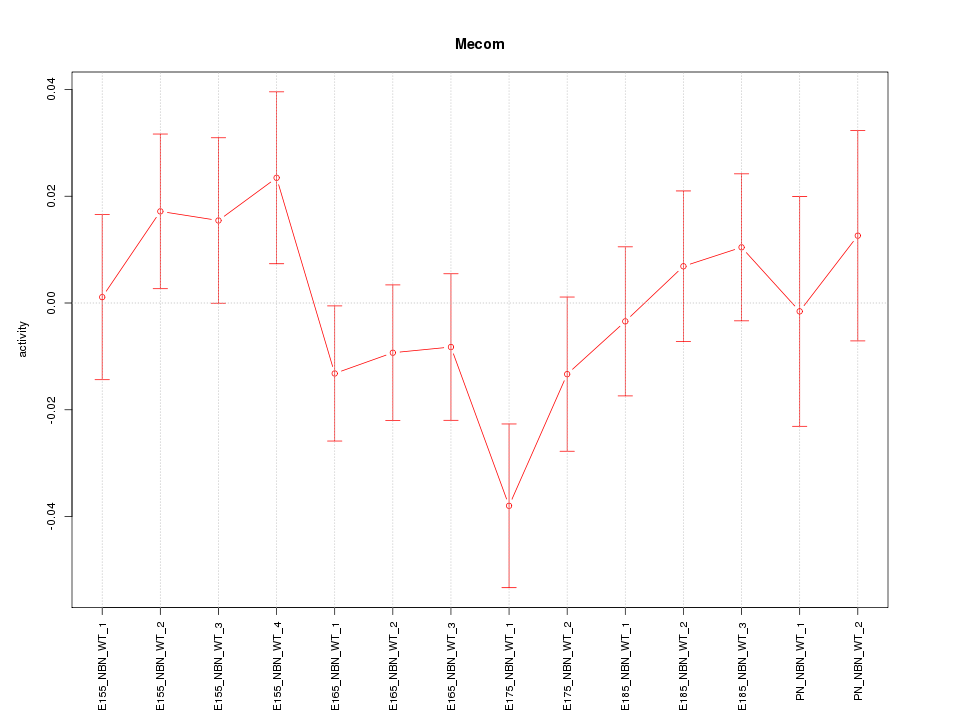
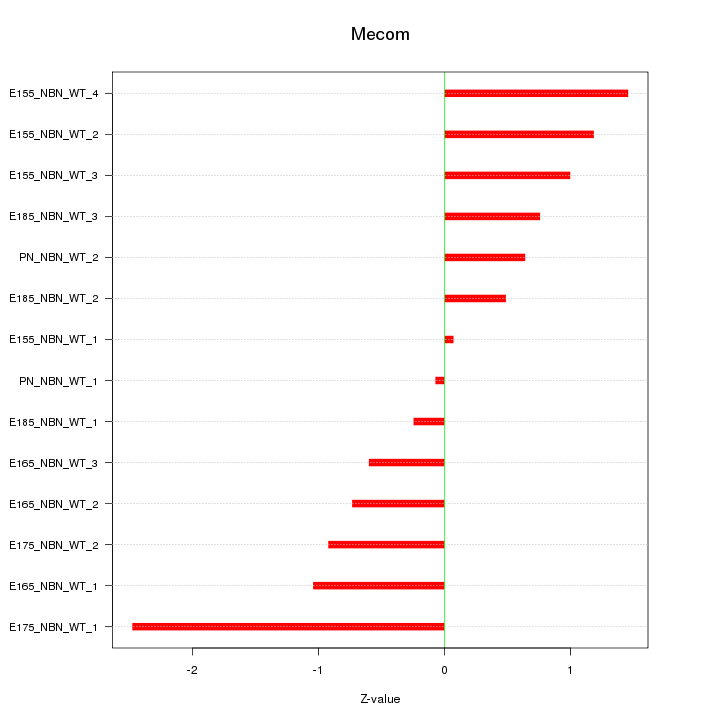
| 2.2 |
15.2 |
GO:0032453 |
histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.4 |
1.6 |
GO:0031721 |
hemoglobin alpha binding(GO:0031721) |
| 0.1 |
4.2 |
GO:0035250 |
UDP-galactosyltransferase activity(GO:0035250) |
| 0.1 |
0.8 |
GO:0048407 |
platelet-derived growth factor binding(GO:0048407) |
| 0.1 |
1.1 |
GO:0004602 |
glutathione peroxidase activity(GO:0004602) |
| 0.0 |
1.3 |
GO:0005385 |
zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 |
0.6 |
GO:0005092 |
GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 |
0.1 |
GO:0003941 |
L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 |
0.1 |
GO:0034714 |
type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 |
0.7 |
GO:0017091 |
AU-rich element binding(GO:0017091) |
| 0.0 |
0.2 |
GO:0003689 |
DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 |
0.3 |
GO:0005540 |
hyaluronic acid binding(GO:0005540) |
| 0.0 |
1.4 |
GO:0043851 |
rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) rRNA (uridine-2'-O-)-methyltransferase activity(GO:0008650) rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) selenocysteine methyltransferase activity(GO:0016205) rRNA (adenine) methyltransferase activity(GO:0016433) rRNA (cytosine) methyltransferase activity(GO:0016434) rRNA (guanine) methyltransferase activity(GO:0016435) 1-phenanthrol methyltransferase activity(GO:0018707) protein-arginine N5-methyltransferase activity(GO:0019702) dimethylarsinite methyltransferase activity(GO:0034541) 4,5-dihydroxybenzo(a)pyrene methyltransferase activity(GO:0034807) 1-hydroxypyrene methyltransferase activity(GO:0034931) 1-hydroxy-6-methoxypyrene methyltransferase activity(GO:0034933) demethylmenaquinone methyltransferase activity(GO:0043770) cobalt-precorrin-6B C5-methyltransferase activity(GO:0043776) cobalt-precorrin-7 C15-methyltransferase activity(GO:0043777) cobalt-precorrin-5B C1-methyltransferase activity(GO:0043780) cobalt-precorrin-3 C17-methyltransferase activity(GO:0043782) dimethylamine methyltransferase activity(GO:0043791) hydroxyneurosporene-O-methyltransferase activity(GO:0043803) tRNA (adenine-57, 58-N(1)-) methyltransferase activity(GO:0043827) methylamine-specific methylcobalamin:coenzyme M methyltransferase activity(GO:0043833) trimethylamine methyltransferase activity(GO:0043834) methanol-specific methylcobalamin:coenzyme M methyltransferase activity(GO:0043851) monomethylamine methyltransferase activity(GO:0043852) P-methyltransferase activity(GO:0051994) Se-methyltransferase activity(GO:0051995) 2-phytyl-1,4-naphthoquinone methyltransferase activity(GO:0052624) tRNA (uracil-2'-O-)-methyltransferase activity(GO:0052665) tRNA (cytosine-2'-O-)-methyltransferase activity(GO:0052666) phosphomethylethanolamine N-methyltransferase activity(GO:0052667) tRNA (cytosine-3-)-methyltransferase activity(GO:0052735) rRNA (cytosine-2'-O-)-methyltransferase activity(GO:0070677) rRNA (cytosine-N4-)-methyltransferase activity(GO:0071424) trihydroxyferuloyl spermidine O-methyltransferase activity(GO:0080012) |