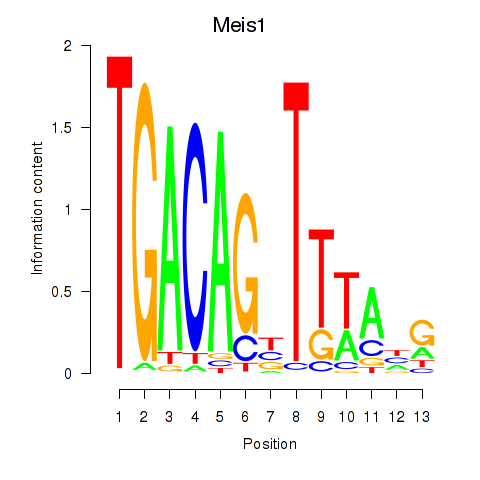
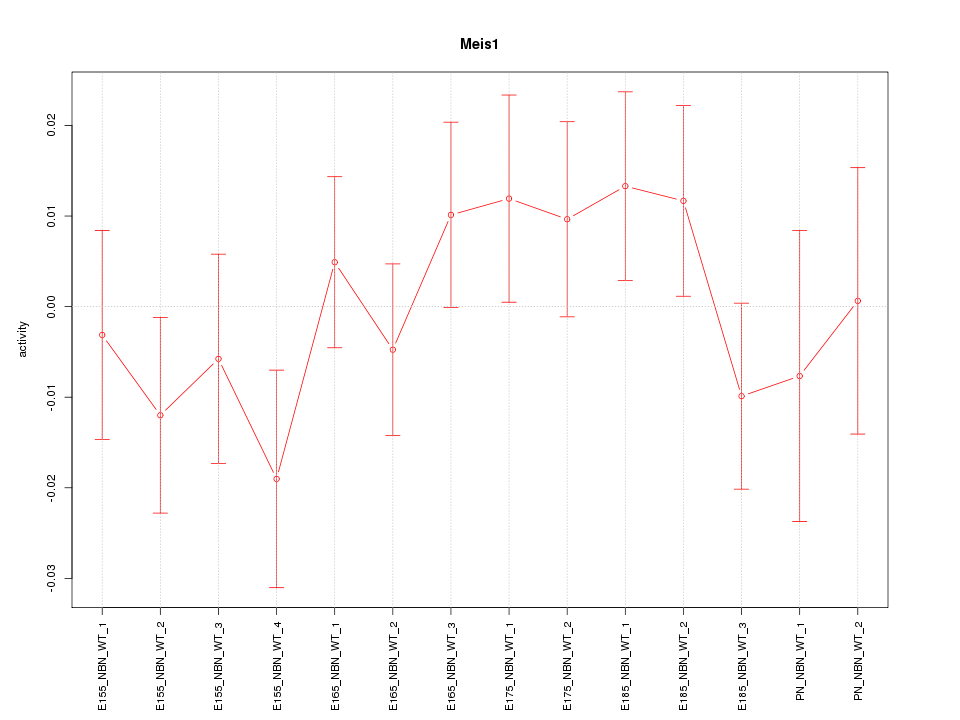
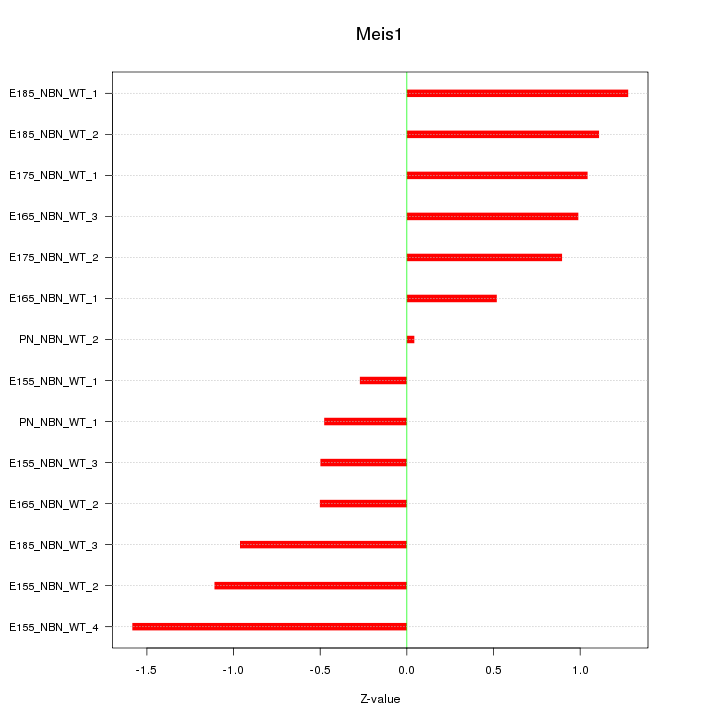
| 0.9 |
2.7 |
GO:0072092 |
ureteric bud invasion(GO:0072092) |
| 0.5 |
3.2 |
GO:0045650 |
negative regulation of macrophage differentiation(GO:0045650) |
| 0.4 |
1.2 |
GO:1901228 |
regulation of osteoclast proliferation(GO:0090289) negative regulation of bone mineralization involved in bone maturation(GO:1900158) positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.3 |
1.9 |
GO:0003383 |
apical constriction(GO:0003383) |
| 0.3 |
0.9 |
GO:0014738 |
regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.3 |
0.9 |
GO:1904395 |
regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.2 |
1.1 |
GO:2000672 |
negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.2 |
1.1 |
GO:2000664 |
positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.2 |
1.2 |
GO:0070474 |
positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.1 |
1.2 |
GO:0071420 |
cellular response to histamine(GO:0071420) |
| 0.1 |
0.5 |
GO:0035609 |
C-terminal protein deglutamylation(GO:0035609) |
| 0.1 |
0.5 |
GO:0015793 |
glycerol transport(GO:0015793) |
| 0.1 |
0.3 |
GO:0046168 |
glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.1 |
0.3 |
GO:1900365 |
positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.1 |
0.3 |
GO:0010808 |
positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.1 |
0.2 |
GO:0060126 |
somatotropin secreting cell differentiation(GO:0060126) |
| 0.1 |
0.2 |
GO:0007621 |
negative regulation of female receptivity(GO:0007621) |
| 0.1 |
0.4 |
GO:1901164 |
negative regulation of trophoblast cell migration(GO:1901164) |
| 0.1 |
0.5 |
GO:0045059 |
positive thymic T cell selection(GO:0045059) |
| 0.1 |
0.6 |
GO:0045719 |
negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.1 |
0.4 |
GO:0045200 |
establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 |
0.4 |
GO:0060018 |
astrocyte fate commitment(GO:0060018) |
| 0.1 |
0.3 |
GO:0090168 |
Golgi reassembly(GO:0090168) |
| 0.1 |
0.7 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 |
0.8 |
GO:0007190 |
activation of adenylate cyclase activity(GO:0007190) |
| 0.0 |
0.3 |
GO:0043504 |
mitochondrial DNA repair(GO:0043504) |
| 0.0 |
0.1 |
GO:0072425 |
signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.0 |
0.4 |
GO:0060294 |
cilium movement involved in cell motility(GO:0060294) |
| 0.0 |
0.1 |
GO:1990481 |
mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 |
0.1 |
GO:0019043 |
positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) establishment of viral latency(GO:0019043) |
| 0.0 |
1.0 |
GO:0007205 |
protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 |
0.1 |
GO:2000642 |
negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.0 |
0.1 |
GO:0050787 |
glycolate metabolic process(GO:0009441) enzyme active site formation via L-cysteine sulfinic acid(GO:0018323) primary alcohol biosynthetic process(GO:0034309) cellular response to glyoxal(GO:0036471) glycolate biosynthetic process(GO:0046295) detoxification of mercury ion(GO:0050787) negative regulation of TRAIL-activated apoptotic signaling pathway(GO:1903122) regulation of pyrroline-5-carboxylate reductase activity(GO:1903167) positive regulation of pyrroline-5-carboxylate reductase activity(GO:1903168) regulation of tyrosine 3-monooxygenase activity(GO:1903176) positive regulation of tyrosine 3-monooxygenase activity(GO:1903178) L-dopa metabolic process(GO:1903184) L-dopa biosynthetic process(GO:1903185) glyoxal metabolic process(GO:1903189) glyoxal catabolic process(GO:1903190) regulation of L-dopa biosynthetic process(GO:1903195) positive regulation of L-dopa biosynthetic process(GO:1903197) regulation of L-dopa decarboxylase activity(GO:1903198) positive regulation of L-dopa decarboxylase activity(GO:1903200) positive regulation of cellular amino acid biosynthetic process(GO:2000284) positive regulation of androgen receptor activity(GO:2000825) |
| 0.0 |
0.2 |
GO:0036438 |
maintenance of lens transparency(GO:0036438) |
| 0.0 |
0.2 |
GO:0034196 |
acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 |
0.1 |
GO:0001951 |
intestinal D-glucose absorption(GO:0001951) terminal web assembly(GO:1902896) |
| 0.0 |
0.1 |
GO:0032289 |
central nervous system myelin formation(GO:0032289) cardiac cell fate specification(GO:0060912) |
| 0.0 |
0.6 |
GO:0001504 |
neurotransmitter uptake(GO:0001504) |
| 0.0 |
0.1 |
GO:0060050 |
positive regulation of protein glycosylation(GO:0060050) |
| 0.0 |
0.3 |
GO:0009143 |
nucleoside triphosphate catabolic process(GO:0009143) |
| 0.0 |
0.1 |
GO:0070126 |
mitochondrial translational termination(GO:0070126) |
| 0.0 |
0.5 |
GO:0001886 |
endothelial cell morphogenesis(GO:0001886) |
| 0.0 |
0.1 |
GO:0052428 |
modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.0 |
0.1 |
GO:1902990 |
mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.0 |
0.2 |
GO:0003062 |
regulation of heart rate by chemical signal(GO:0003062) |
| 0.0 |
0.1 |
GO:0044034 |
negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 |
0.1 |
GO:0034316 |
negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 |
0.1 |
GO:0030049 |
muscle filament sliding(GO:0030049) |
| 0.0 |
0.1 |
GO:0044375 |
regulation of peroxisome size(GO:0044375) |
| 0.0 |
0.3 |
GO:0060363 |
cranial suture morphogenesis(GO:0060363) |
| 0.0 |
0.2 |
GO:0042573 |
retinoic acid metabolic process(GO:0042573) |
| 0.0 |
0.1 |
GO:0048478 |
replication fork protection(GO:0048478) |
| 0.0 |
0.5 |
GO:0046827 |
positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 |
0.0 |
GO:0042271 |
susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 |
0.1 |
GO:1904262 |
negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 |
0.2 |
GO:0050909 |
sensory perception of taste(GO:0050909) |
| 0.0 |
0.5 |
GO:0031146 |
SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 |
0.2 |
GO:0010758 |
regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 |
0.3 |
GO:0006182 |
cGMP biosynthetic process(GO:0006182) |
| 0.0 |
0.3 |
GO:0045475 |
locomotor rhythm(GO:0045475) |
| 0.0 |
0.1 |
GO:0060999 |
positive regulation of dendritic spine development(GO:0060999) |
| 0.0 |
0.0 |
GO:0046671 |
negative regulation of retinal cell programmed cell death(GO:0046671) |