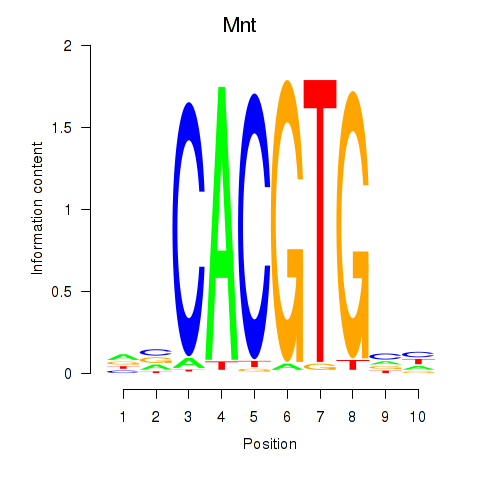
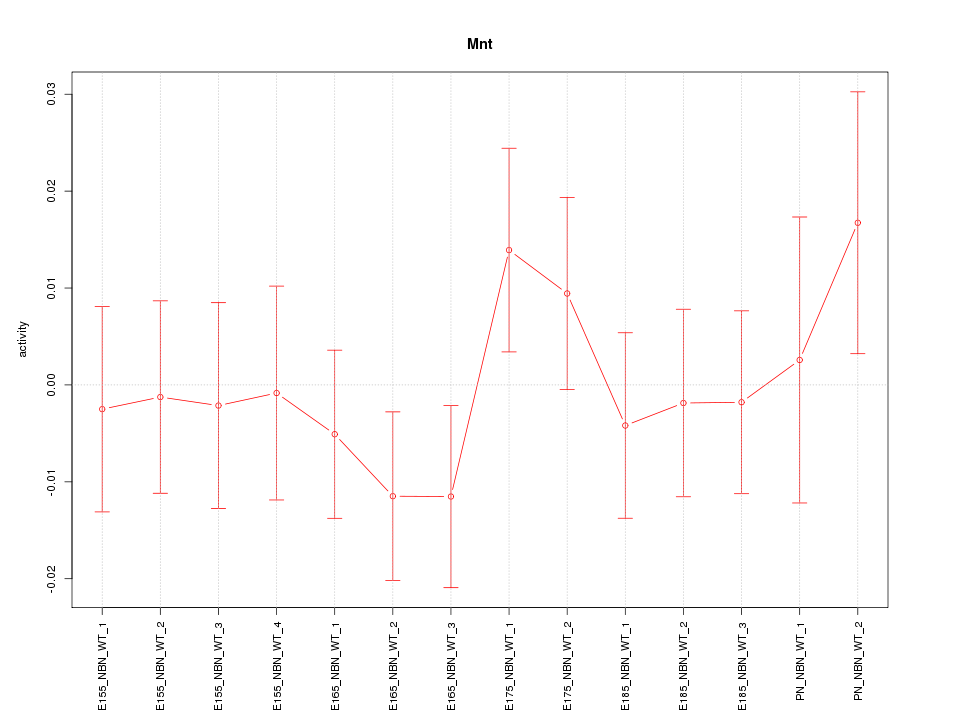
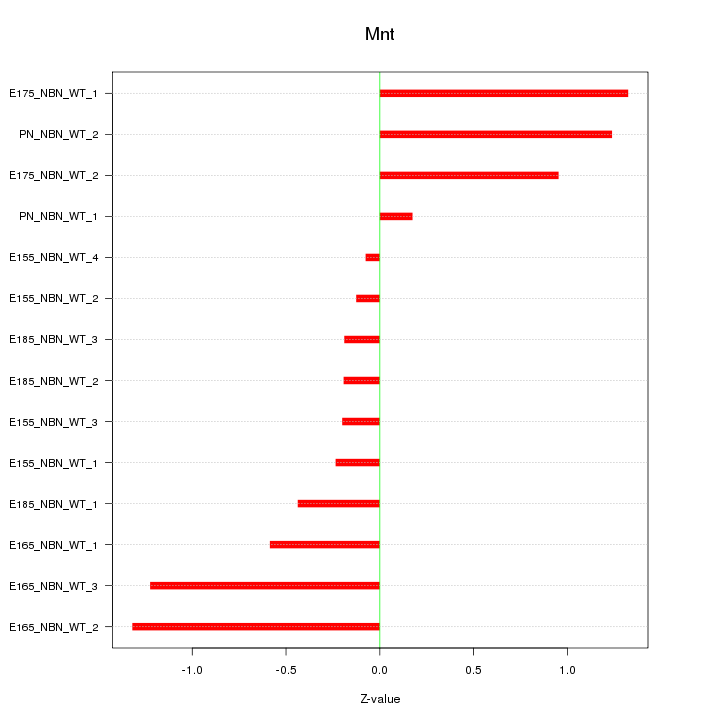
| 0.6 |
1.7 |
GO:0003349 |
epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.5 |
2.6 |
GO:1904426 |
positive regulation of GTP binding(GO:1904426) |
| 0.5 |
1.5 |
GO:0072034 |
renal vesicle induction(GO:0072034) |
| 0.4 |
3.7 |
GO:0060665 |
regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.4 |
1.1 |
GO:0010845 |
positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.3 |
0.7 |
GO:0060437 |
lung growth(GO:0060437) |
| 0.3 |
1.0 |
GO:0021893 |
cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 0.3 |
1.6 |
GO:0097116 |
gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.3 |
0.9 |
GO:0060729 |
intestinal epithelial structure maintenance(GO:0060729) |
| 0.3 |
0.9 |
GO:1904685 |
positive regulation of metalloendopeptidase activity(GO:1904685) |
| 0.3 |
0.9 |
GO:1902358 |
sulfate transmembrane transport(GO:1902358) |
| 0.3 |
0.8 |
GO:0060023 |
soft palate development(GO:0060023) |
| 0.3 |
2.2 |
GO:0071557 |
histone H3-K27 demethylation(GO:0071557) |
| 0.3 |
1.3 |
GO:0070460 |
thyroid-stimulating hormone secretion(GO:0070460) |
| 0.3 |
1.0 |
GO:0007066 |
female meiosis sister chromatid cohesion(GO:0007066) |
| 0.2 |
0.7 |
GO:1904706 |
response to cisplatin(GO:0072718) negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.2 |
0.7 |
GO:0001732 |
formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.2 |
2.2 |
GO:0051901 |
positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.2 |
0.7 |
GO:0035701 |
hematopoietic stem cell migration(GO:0035701) |
| 0.2 |
0.2 |
GO:0045901 |
positive regulation of translational elongation(GO:0045901) |
| 0.2 |
0.6 |
GO:1904049 |
regulation of spontaneous neurotransmitter secretion(GO:1904048) negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.2 |
0.6 |
GO:0009177 |
deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) |
| 0.2 |
0.8 |
GO:0001907 |
killing by symbiont of host cells(GO:0001907) induction of programmed cell death(GO:0012502) disruption by symbiont of host cell(GO:0044004) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) positive regulation of apoptotic process by virus(GO:0060139) positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.2 |
1.1 |
GO:0033600 |
negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.2 |
0.3 |
GO:0032423 |
regulation of mismatch repair(GO:0032423) |
| 0.2 |
1.0 |
GO:0048254 |
snoRNA localization(GO:0048254) |
| 0.2 |
0.5 |
GO:0044340 |
canonical Wnt signaling pathway involved in regulation of cell proliferation(GO:0044340) |
| 0.2 |
0.9 |
GO:0003383 |
apical constriction(GO:0003383) |
| 0.2 |
0.6 |
GO:0010891 |
negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.2 |
0.8 |
GO:0050847 |
progesterone receptor signaling pathway(GO:0050847) |
| 0.2 |
0.5 |
GO:0002842 |
T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) positive regulation of T cell mediated immune response to tumor cell(GO:0002842) |
| 0.1 |
0.6 |
GO:0055099 |
regulation of Cdc42 protein signal transduction(GO:0032489) response to high density lipoprotein particle(GO:0055099) platelet dense granule organization(GO:0060155) |
| 0.1 |
0.4 |
GO:0015712 |
hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 |
1.0 |
GO:0006685 |
sphingomyelin catabolic process(GO:0006685) |
| 0.1 |
0.6 |
GO:1990928 |
response to amino acid starvation(GO:1990928) |
| 0.1 |
0.7 |
GO:0015015 |
heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 |
0.4 |
GO:0042822 |
vitamin B6 metabolic process(GO:0042816) pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.1 |
0.4 |
GO:0014738 |
regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.1 |
0.5 |
GO:0035752 |
lysosomal lumen pH elevation(GO:0035752) |
| 0.1 |
0.3 |
GO:0021759 |
globus pallidus development(GO:0021759) |
| 0.1 |
0.7 |
GO:0000050 |
urea cycle(GO:0000050) urea metabolic process(GO:0019627) |
| 0.1 |
0.9 |
GO:2000973 |
regulation of pro-B cell differentiation(GO:2000973) |
| 0.1 |
0.3 |
GO:1903416 |
response to glycoside(GO:1903416) |
| 0.1 |
0.7 |
GO:0070601 |
centromeric sister chromatid cohesion(GO:0070601) |
| 0.1 |
0.7 |
GO:0042518 |
negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.1 |
0.5 |
GO:2000680 |
regulation of rubidium ion transport(GO:2000680) |
| 0.1 |
0.8 |
GO:0006265 |
DNA topological change(GO:0006265) |
| 0.1 |
0.4 |
GO:1902626 |
assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 |
1.0 |
GO:0070574 |
cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 |
0.9 |
GO:0002087 |
regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.1 |
0.7 |
GO:0008616 |
queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.1 |
0.5 |
GO:0006543 |
glutamine catabolic process(GO:0006543) |
| 0.1 |
1.0 |
GO:0003215 |
cardiac right ventricle morphogenesis(GO:0003215) |
| 0.1 |
0.4 |
GO:0046487 |
glyoxylate metabolic process(GO:0046487) |
| 0.1 |
0.6 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 |
0.4 |
GO:0090467 |
L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) |
| 0.1 |
0.3 |
GO:0046900 |
tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.1 |
2.5 |
GO:0043153 |
entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 |
0.4 |
GO:0045347 |
negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.1 |
2.1 |
GO:2000310 |
regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.1 |
0.8 |
GO:0035520 |
monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.1 |
0.6 |
GO:0000395 |
mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 |
0.2 |
GO:0060399 |
positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.1 |
0.3 |
GO:0072015 |
glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.1 |
0.4 |
GO:0032346 |
positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 0.1 |
0.8 |
GO:0061158 |
3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 |
0.2 |
GO:0045113 |
regulation of integrin biosynthetic process(GO:0045113) |
| 0.1 |
0.2 |
GO:0014877 |
response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 |
0.3 |
GO:0071930 |
negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.1 |
0.8 |
GO:0097154 |
GABAergic neuron differentiation(GO:0097154) |
| 0.1 |
0.3 |
GO:0048633 |
positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.1 |
0.3 |
GO:0048312 |
intracellular distribution of mitochondria(GO:0048312) |
| 0.1 |
0.2 |
GO:0035526 |
retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 |
0.8 |
GO:0032688 |
negative regulation of interferon-beta production(GO:0032688) |
| 0.1 |
0.2 |
GO:0007161 |
calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.1 |
0.2 |
GO:0098528 |
skeletal muscle fiber differentiation(GO:0098528) |
| 0.1 |
0.6 |
GO:0032876 |
negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 |
0.2 |
GO:2000015 |
regulation of determination of dorsal identity(GO:2000015) |
| 0.1 |
1.2 |
GO:0021859 |
pyramidal neuron differentiation(GO:0021859) |
| 0.1 |
0.6 |
GO:0006273 |
lagging strand elongation(GO:0006273) |
| 0.1 |
0.3 |
GO:2000189 |
positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 |
0.3 |
GO:0046477 |
glycosylceramide catabolic process(GO:0046477) |
| 0.1 |
0.2 |
GO:0070536 |
protein K63-linked deubiquitination(GO:0070536) |
| 0.1 |
0.1 |
GO:0060025 |
regulation of synaptic activity(GO:0060025) |
| 0.1 |
0.3 |
GO:0045053 |
protein retention in Golgi apparatus(GO:0045053) |
| 0.1 |
0.5 |
GO:0000083 |
regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 |
0.4 |
GO:1903441 |
protein localization to ciliary membrane(GO:1903441) |
| 0.1 |
0.4 |
GO:2000394 |
positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.1 |
0.2 |
GO:0019919 |
peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.1 |
0.6 |
GO:0090557 |
establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 |
0.4 |
GO:0006983 |
ER overload response(GO:0006983) |
| 0.1 |
0.1 |
GO:0034723 |
DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 |
0.8 |
GO:0002021 |
response to dietary excess(GO:0002021) |
| 0.1 |
0.3 |
GO:0016080 |
synaptic vesicle targeting(GO:0016080) protein localization to adherens junction(GO:0071896) |
| 0.1 |
0.2 |
GO:0019276 |
UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 |
0.3 |
GO:0090209 |
negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.0 |
1.0 |
GO:0034453 |
microtubule anchoring(GO:0034453) |
| 0.0 |
0.1 |
GO:0045350 |
interferon-beta biosynthetic process(GO:0045350) regulation of interferon-beta biosynthetic process(GO:0045357) positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.0 |
0.2 |
GO:0071442 |
positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 |
0.3 |
GO:1903071 |
positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 |
0.2 |
GO:2000253 |
positive regulation of feeding behavior(GO:2000253) |
| 0.0 |
0.1 |
GO:0035964 |
COPI-coated vesicle budding(GO:0035964) |
| 0.0 |
0.2 |
GO:0061428 |
embryonic heart tube left/right pattern formation(GO:0060971) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.0 |
0.4 |
GO:0061088 |
sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 |
0.7 |
GO:0007130 |
synaptonemal complex assembly(GO:0007130) |
| 0.0 |
0.3 |
GO:0010747 |
positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.0 |
0.3 |
GO:1901970 |
meiotic chromosome separation(GO:0051307) positive regulation of mitotic sister chromatid separation(GO:1901970) |
| 0.0 |
0.6 |
GO:1904816 |
positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.0 |
0.3 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.0 |
0.2 |
GO:0033700 |
phospholipid efflux(GO:0033700) |
| 0.0 |
0.1 |
GO:1904569 |
regulation of selenocysteine incorporation(GO:1904569) |
| 0.0 |
0.3 |
GO:0015862 |
uridine transport(GO:0015862) |
| 0.0 |
0.6 |
GO:0043402 |
glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 |
0.2 |
GO:1900246 |
positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 |
0.3 |
GO:0070734 |
histone H3-K27 methylation(GO:0070734) |
| 0.0 |
1.1 |
GO:0045070 |
positive regulation of viral genome replication(GO:0045070) |
| 0.0 |
0.1 |
GO:0034162 |
toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.0 |
0.1 |
GO:0009174 |
'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) UMP biosynthetic process(GO:0006222) pyrimidine nucleoside monophosphate biosynthetic process(GO:0009130) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) pyrimidine nucleobase biosynthetic process(GO:0019856) UMP metabolic process(GO:0046049) |
| 0.0 |
0.5 |
GO:0021942 |
radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 |
0.1 |
GO:0070889 |
platelet alpha granule organization(GO:0070889) |
| 0.0 |
0.1 |
GO:0035356 |
cellular triglyceride homeostasis(GO:0035356) |
| 0.0 |
0.4 |
GO:0060689 |
cell differentiation involved in salivary gland development(GO:0060689) |
| 0.0 |
0.1 |
GO:0034975 |
protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 |
0.7 |
GO:0071786 |
endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 |
0.1 |
GO:2000211 |
regulation of glutamate metabolic process(GO:2000211) |
| 0.0 |
0.4 |
GO:2000505 |
regulation of energy homeostasis(GO:2000505) |
| 0.0 |
0.2 |
GO:0090148 |
membrane fission(GO:0090148) |
| 0.0 |
0.1 |
GO:0019509 |
L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 |
1.0 |
GO:0048255 |
mRNA stabilization(GO:0048255) |
| 0.0 |
0.2 |
GO:1903301 |
positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 |
0.3 |
GO:0007141 |
male meiosis I(GO:0007141) |
| 0.0 |
0.4 |
GO:0097186 |
amelogenesis(GO:0097186) |
| 0.0 |
0.8 |
GO:0007638 |
mechanosensory behavior(GO:0007638) |
| 0.0 |
0.1 |
GO:0032053 |
ciliary basal body organization(GO:0032053) |
| 0.0 |
0.1 |
GO:2000304 |
positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 |
0.4 |
GO:0015937 |
coenzyme A biosynthetic process(GO:0015937) |
| 0.0 |
0.4 |
GO:0071281 |
cellular response to iron ion(GO:0071281) |
| 0.0 |
0.1 |
GO:0042360 |
vitamin E metabolic process(GO:0042360) intermembrane transport(GO:0046909) |
| 0.0 |
0.2 |
GO:0006855 |
drug transmembrane transport(GO:0006855) |
| 0.0 |
0.1 |
GO:0043988 |
histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 |
0.1 |
GO:0006011 |
UDP-glucose metabolic process(GO:0006011) |
| 0.0 |
0.4 |
GO:0051457 |
maintenance of protein location in nucleus(GO:0051457) |
| 0.0 |
0.2 |
GO:1901409 |
regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 |
0.3 |
GO:0030322 |
stabilization of membrane potential(GO:0030322) |
| 0.0 |
0.1 |
GO:0051182 |
coenzyme transport(GO:0051182) |
| 0.0 |
0.1 |
GO:0048873 |
homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 |
0.1 |
GO:1902369 |
negative regulation of RNA catabolic process(GO:1902369) |
| 0.0 |
0.1 |
GO:2000049 |
positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 |
0.1 |
GO:0045006 |
DNA deamination(GO:0045006) |
| 0.0 |
0.3 |
GO:1990126 |
retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 |
0.8 |
GO:0006270 |
DNA replication initiation(GO:0006270) |
| 0.0 |
0.1 |
GO:0051503 |
adenine nucleotide transport(GO:0051503) |
| 0.0 |
0.1 |
GO:2000045 |
regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.0 |
0.8 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.0 |
0.4 |
GO:0032933 |
response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 |
0.1 |
GO:0042373 |
vitamin K metabolic process(GO:0042373) |
| 0.0 |
0.2 |
GO:0035542 |
regulation of SNARE complex assembly(GO:0035542) |
| 0.0 |
0.1 |
GO:0007253 |
cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 |
1.0 |
GO:0007628 |
adult walking behavior(GO:0007628) walking behavior(GO:0090659) |
| 0.0 |
0.2 |
GO:0060213 |
regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 |
0.5 |
GO:0034198 |
cellular response to amino acid starvation(GO:0034198) |
| 0.0 |
0.0 |
GO:0007386 |
compartment pattern specification(GO:0007386) |
| 0.0 |
0.1 |
GO:0051784 |
negative regulation of mitotic nuclear division(GO:0045839) negative regulation of nuclear division(GO:0051784) |
| 0.0 |
0.2 |
GO:1904153 |
negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 |
0.1 |
GO:0089711 |
L-glutamate transmembrane transport(GO:0089711) |
| 0.0 |
0.2 |
GO:0006308 |
DNA catabolic process(GO:0006308) |
| 0.0 |
0.8 |
GO:0000281 |
mitotic cytokinesis(GO:0000281) |
| 0.0 |
0.2 |
GO:0033089 |
positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 |
0.8 |
GO:0071277 |
cellular response to calcium ion(GO:0071277) |
| 0.0 |
0.1 |
GO:0034316 |
negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 |
0.2 |
GO:0034143 |
regulation of toll-like receptor 4 signaling pathway(GO:0034143) |
| 0.0 |
0.1 |
GO:0099515 |
actin filament-based transport(GO:0099515) |
| 0.0 |
0.1 |
GO:1900028 |
negative regulation of ruffle assembly(GO:1900028) |
| 0.0 |
0.5 |
GO:0001709 |
cell fate determination(GO:0001709) |
| 0.0 |
0.0 |
GO:0032877 |
positive regulation of DNA endoreduplication(GO:0032877) |
| 0.0 |
0.3 |
GO:0000245 |
spliceosomal complex assembly(GO:0000245) |
| 0.0 |
0.1 |
GO:0032211 |
negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.0 |
0.7 |
GO:0046329 |
negative regulation of JNK cascade(GO:0046329) |
| 0.0 |
0.1 |
GO:0045717 |
negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 |
0.4 |
GO:0018345 |
protein palmitoylation(GO:0018345) |
| 0.0 |
0.1 |
GO:0070358 |
actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 |
0.2 |
GO:0000244 |
spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 |
0.0 |
GO:0019074 |
viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 |
0.3 |
GO:0030201 |
heparan sulfate proteoglycan metabolic process(GO:0030201) |
| 0.0 |
0.5 |
GO:0045022 |
early endosome to late endosome transport(GO:0045022) |
| 0.0 |
0.1 |
GO:0043461 |
proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 |
0.2 |
GO:0043374 |
CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 |
0.3 |
GO:0001921 |
positive regulation of receptor recycling(GO:0001921) |
| 0.0 |
0.3 |
GO:0021522 |
spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 |
0.1 |
GO:0035721 |
intraciliary retrograde transport(GO:0035721) |
| 0.0 |
0.5 |
GO:0035019 |
somatic stem cell population maintenance(GO:0035019) |
| 0.0 |
0.4 |
GO:0070830 |
bicellular tight junction assembly(GO:0070830) |
| 0.0 |
0.1 |
GO:0006390 |
transcription from mitochondrial promoter(GO:0006390) |
| 0.0 |
0.0 |
GO:0045039 |
protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 |
0.7 |
GO:0030500 |
regulation of bone mineralization(GO:0030500) |
| 0.0 |
0.0 |
GO:0003278 |
apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.0 |
0.0 |
GO:0008295 |
spermidine biosynthetic process(GO:0008295) |
| 0.0 |
0.0 |
GO:0006545 |
glycine biosynthetic process(GO:0006545) |
| 0.0 |
0.2 |
GO:0006388 |
tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 |
0.0 |
GO:0006592 |
ornithine biosynthetic process(GO:0006592) |
| 0.0 |
0.2 |
GO:0006515 |
misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 |
1.5 |
GO:0010771 |
negative regulation of cell morphogenesis involved in differentiation(GO:0010771) |
| 0.0 |
0.1 |
GO:0002566 |
somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 |
0.2 |
GO:0046548 |
retinal rod cell development(GO:0046548) |
| 0.0 |
0.1 |
GO:0010388 |
protein deneddylation(GO:0000338) cullin deneddylation(GO:0010388) |
| 0.0 |
0.1 |
GO:0008355 |
olfactory learning(GO:0008355) |