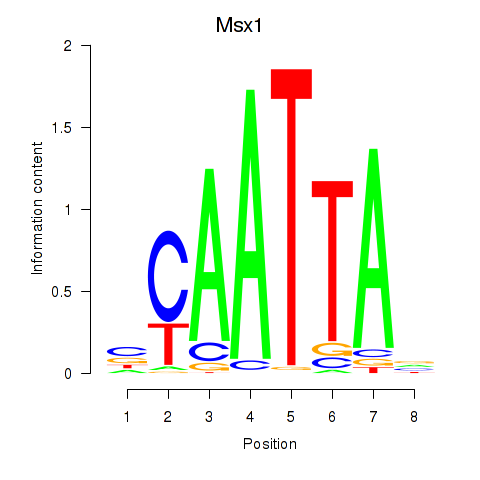
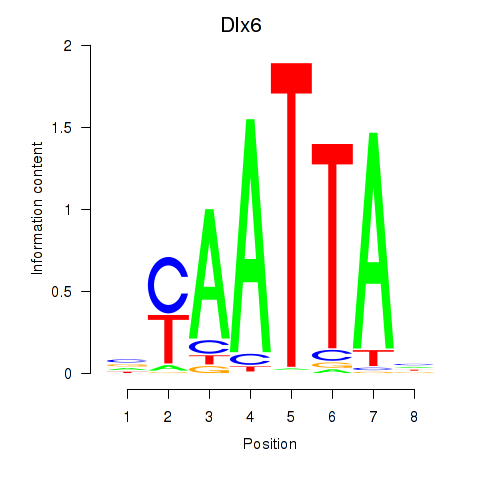
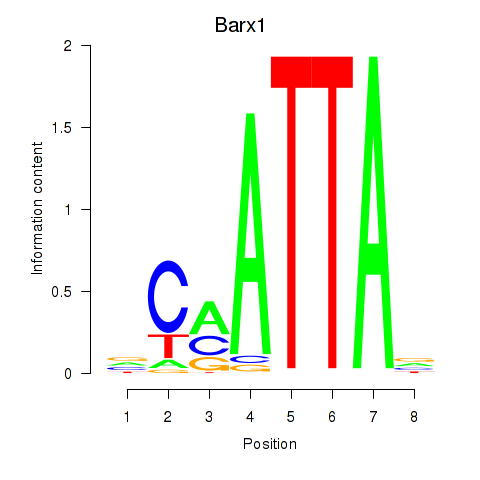
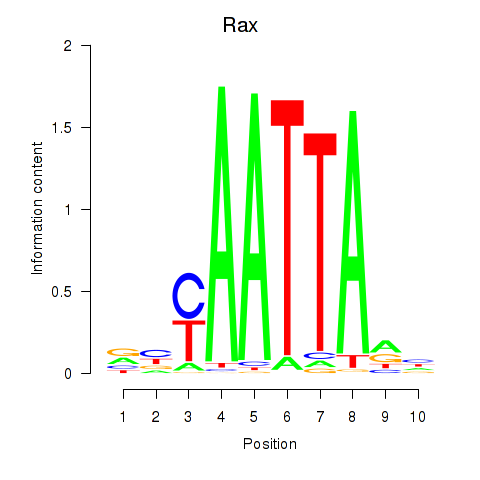
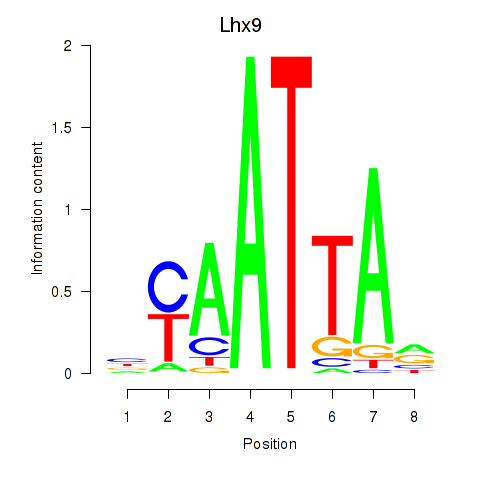
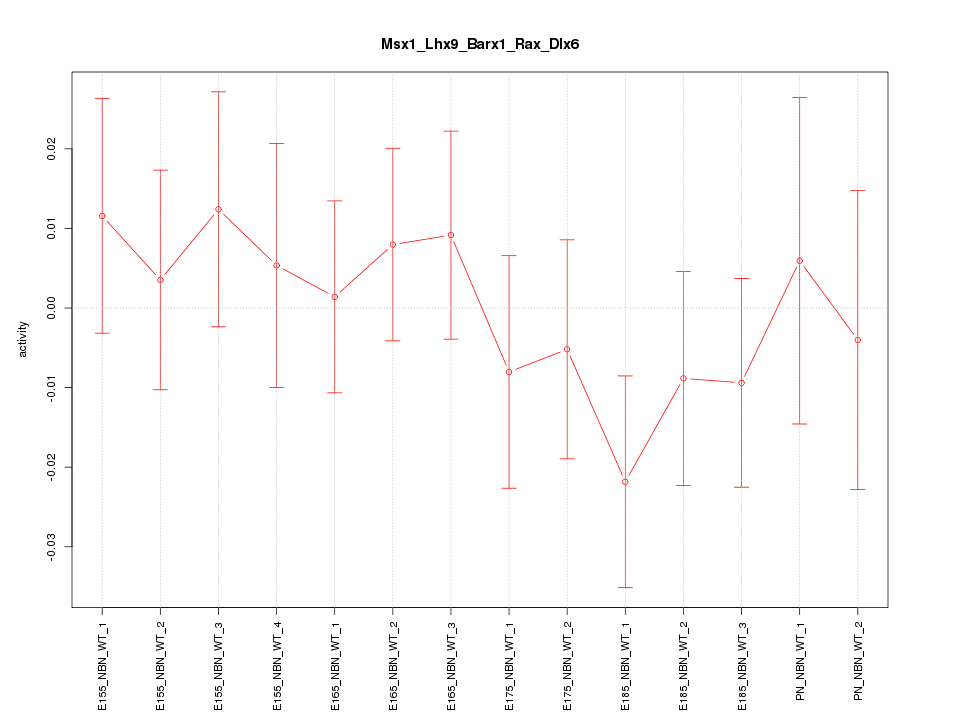
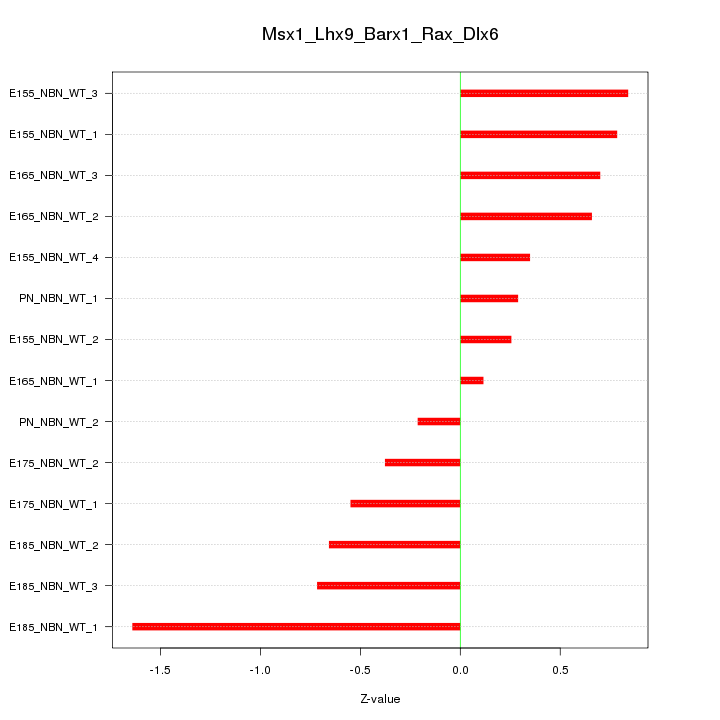
| 4.5 |
13.6 |
GO:0002302 |
CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.8 |
2.5 |
GO:0042663 |
regulation of endodermal cell fate specification(GO:0042663) |
| 0.8 |
5.0 |
GO:0030643 |
cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.4 |
2.0 |
GO:0038032 |
termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.3 |
1.6 |
GO:0003150 |
muscular septum morphogenesis(GO:0003150) |
| 0.3 |
0.9 |
GO:0050975 |
sensory perception of touch(GO:0050975) |
| 0.3 |
1.4 |
GO:0048245 |
eosinophil chemotaxis(GO:0048245) |
| 0.2 |
0.7 |
GO:0097155 |
fasciculation of sensory neuron axon(GO:0097155) |
| 0.2 |
1.0 |
GO:0021847 |
ventricular zone neuroblast division(GO:0021847) |
| 0.2 |
0.2 |
GO:0071336 |
fibroblast growth factor receptor signaling pathway involved in mammary gland specification(GO:0060595) mammary gland bud formation(GO:0060615) branch elongation involved in salivary gland morphogenesis(GO:0060667) mesenchymal cell differentiation involved in lung development(GO:0060915) regulation of hair follicle cell proliferation(GO:0071336) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.2 |
2.3 |
GO:0060501 |
positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.2 |
0.7 |
GO:0061055 |
myotome development(GO:0061055) |
| 0.2 |
0.5 |
GO:0021852 |
pyramidal neuron migration(GO:0021852) |
| 0.1 |
0.4 |
GO:0001543 |
ovarian follicle rupture(GO:0001543) |
| 0.1 |
1.0 |
GO:0031536 |
positive regulation of exit from mitosis(GO:0031536) |
| 0.1 |
1.0 |
GO:0035970 |
peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 |
0.5 |
GO:1903056 |
regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.1 |
0.2 |
GO:0060023 |
soft palate development(GO:0060023) |
| 0.1 |
0.5 |
GO:0032625 |
interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 |
0.2 |
GO:0032512 |
regulation of protein phosphatase type 2B activity(GO:0032512) positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.1 |
0.2 |
GO:0035063 |
nuclear speck organization(GO:0035063) |
| 0.1 |
0.4 |
GO:0002326 |
B cell lineage commitment(GO:0002326) |
| 0.1 |
0.2 |
GO:0042271 |
susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 |
0.3 |
GO:0042636 |
negative regulation of hair cycle(GO:0042636) progesterone secretion(GO:0042701) negative regulation of hair follicle development(GO:0051799) positive regulation of ovulation(GO:0060279) |
| 0.0 |
1.1 |
GO:0097352 |
autophagosome maturation(GO:0097352) |
| 0.0 |
0.1 |
GO:0035864 |
response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.0 |
0.4 |
GO:0007195 |
adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 |
1.1 |
GO:0090036 |
regulation of protein kinase C signaling(GO:0090036) |
| 0.0 |
0.2 |
GO:0070562 |
regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 |
0.1 |
GO:1904565 |
response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.0 |
0.1 |
GO:0048819 |
embryonic nail plate morphogenesis(GO:0035880) negative regulation of keratinocyte differentiation(GO:0045617) positive regulation of hair follicle maturation(GO:0048818) regulation of hair follicle maturation(GO:0048819) regulation of catagen(GO:0051794) positive regulation of catagen(GO:0051795) frontal suture morphogenesis(GO:0060364) |
| 0.0 |
0.1 |
GO:0006933 |
negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 |
0.1 |
GO:0001550 |
ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 |
0.7 |
GO:0001574 |
ganglioside biosynthetic process(GO:0001574) |
| 0.0 |
0.6 |
GO:0060218 |
hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 |
0.1 |
GO:0035553 |
oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 |
0.3 |
GO:0061302 |
smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 |
0.5 |
GO:0001833 |
inner cell mass cell proliferation(GO:0001833) |
| 0.0 |
0.5 |
GO:0007530 |
sex determination(GO:0007530) |
| 0.0 |
0.1 |
GO:0000707 |
meiotic DNA recombinase assembly(GO:0000707) resolution of recombination intermediates(GO:0071139) |
| 0.0 |
0.1 |
GO:0006543 |
glutamine catabolic process(GO:0006543) |
| 0.0 |
0.5 |
GO:0048305 |
immunoglobulin secretion(GO:0048305) |
| 0.0 |
0.5 |
GO:0008340 |
determination of adult lifespan(GO:0008340) |
| 0.0 |
0.3 |
GO:0021860 |
pyramidal neuron development(GO:0021860) |
| 0.0 |
0.1 |
GO:0090274 |
positive regulation of somatostatin secretion(GO:0090274) |
| 0.0 |
0.7 |
GO:0003301 |
physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 |
1.0 |
GO:0006284 |
base-excision repair(GO:0006284) |
| 0.0 |
0.1 |
GO:0060060 |
post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.0 |
0.6 |
GO:0009409 |
response to cold(GO:0009409) |
| 0.0 |
0.1 |
GO:0016198 |
axon choice point recognition(GO:0016198) |
| 0.0 |
0.3 |
GO:0014049 |
positive regulation of glutamate secretion(GO:0014049) |
| 0.0 |
0.1 |
GO:0090325 |
regulation of locomotion involved in locomotory behavior(GO:0090325) negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.0 |
0.2 |
GO:0035887 |
aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 |
0.1 |
GO:0042536 |
negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.0 |
0.1 |
GO:0009838 |
abscission(GO:0009838) positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 |
0.1 |
GO:0030327 |
prenylated protein catabolic process(GO:0030327) |
| 0.0 |
0.9 |
GO:0045600 |
positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 |
0.1 |
GO:0031017 |
exocrine pancreas development(GO:0031017) |
| 0.0 |
0.1 |
GO:0034587 |
piRNA metabolic process(GO:0034587) |
| 0.0 |
0.2 |
GO:0060134 |
prepulse inhibition(GO:0060134) |
| 0.0 |
0.0 |
GO:0003278 |
apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.0 |
0.1 |
GO:0016259 |
selenocysteine metabolic process(GO:0016259) |
| 0.0 |
0.1 |
GO:0071578 |
zinc II ion transmembrane import(GO:0071578) |
| 0.0 |
0.3 |
GO:0060074 |
synapse maturation(GO:0060074) |
| 0.0 |
1.3 |
GO:0098792 |
xenophagy(GO:0098792) |
| 0.0 |
4.8 |
GO:0016055 |
Wnt signaling pathway(GO:0016055) cell-cell signaling by wnt(GO:0198738) |
| 0.0 |
0.1 |
GO:0051684 |
maintenance of Golgi location(GO:0051684) |
| 0.0 |
0.1 |
GO:2000741 |
positive regulation of mesenchymal stem cell differentiation(GO:2000741) |