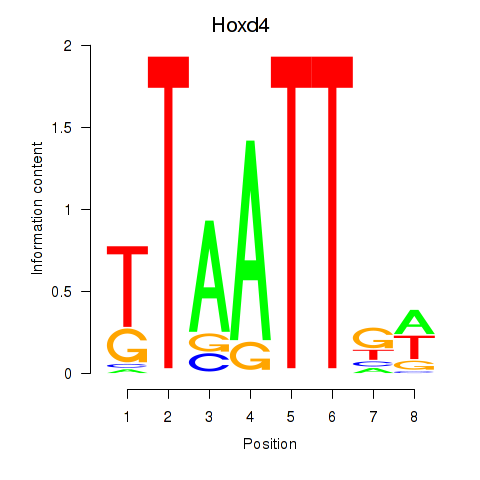
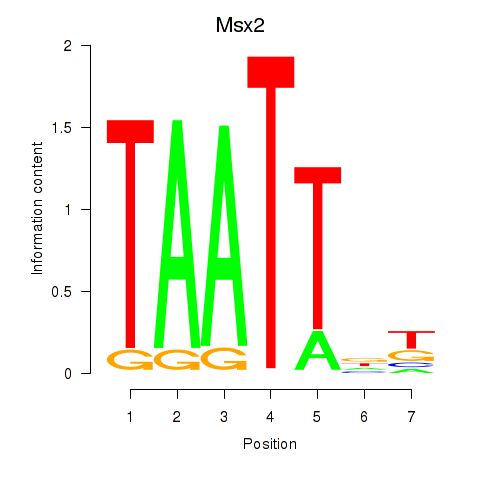
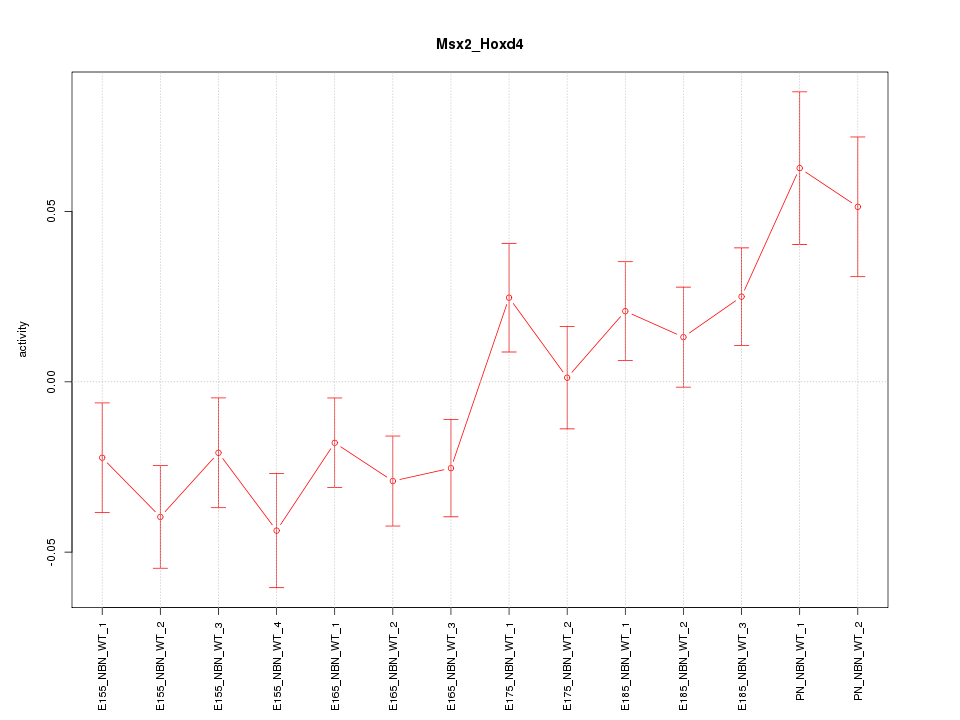
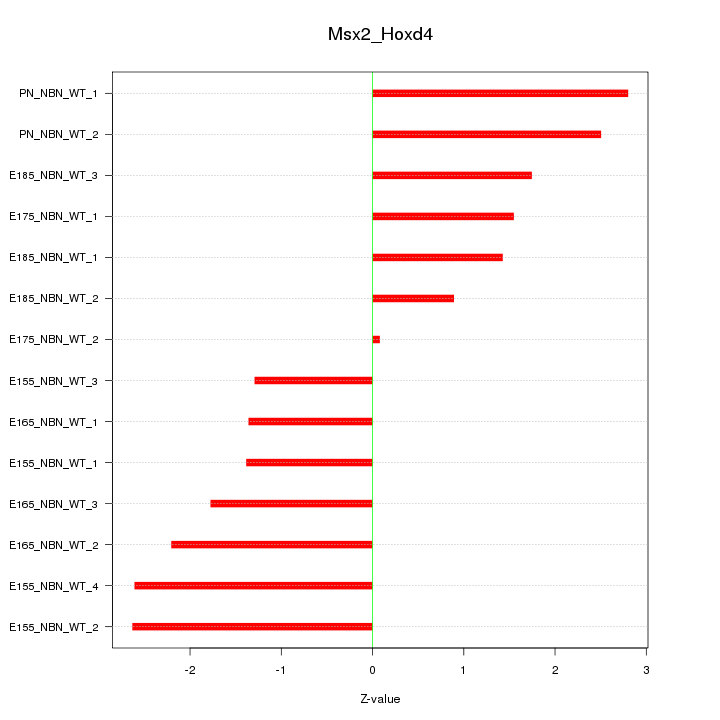
| 7.0 |
21.1 |
GO:0090425 |
hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 1.3 |
3.8 |
GO:1904395 |
regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.5 |
6.8 |
GO:0051764 |
actin crosslink formation(GO:0051764) |
| 0.5 |
1.9 |
GO:1903378 |
positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.4 |
1.2 |
GO:0030827 |
negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.4 |
2.2 |
GO:0060648 |
mammary gland bud morphogenesis(GO:0060648) |
| 0.4 |
1.1 |
GO:0050929 |
corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.3 |
2.7 |
GO:0071732 |
cellular response to nitric oxide(GO:0071732) |
| 0.3 |
1.5 |
GO:0097116 |
gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.3 |
2.4 |
GO:0006620 |
posttranslational protein targeting to membrane(GO:0006620) |
| 0.3 |
1.3 |
GO:0046601 |
positive regulation of centriole replication(GO:0046601) |
| 0.3 |
0.8 |
GO:0006419 |
alanyl-tRNA aminoacylation(GO:0006419) |
| 0.2 |
0.7 |
GO:0035106 |
operant conditioning(GO:0035106) |
| 0.2 |
0.7 |
GO:0030916 |
otic vesicle formation(GO:0030916) |
| 0.2 |
0.7 |
GO:1901420 |
negative regulation of response to alcohol(GO:1901420) |
| 0.2 |
5.0 |
GO:0007638 |
mechanosensory behavior(GO:0007638) |
| 0.2 |
1.7 |
GO:0019511 |
peptidyl-proline hydroxylation(GO:0019511) |
| 0.2 |
3.0 |
GO:0006953 |
acute-phase response(GO:0006953) |
| 0.2 |
2.8 |
GO:0032060 |
bleb assembly(GO:0032060) |
| 0.1 |
5.0 |
GO:0031365 |
N-terminal protein amino acid modification(GO:0031365) |
| 0.1 |
1.0 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.1 |
0.7 |
GO:0070171 |
negative regulation of tooth mineralization(GO:0070171) |
| 0.1 |
2.2 |
GO:0072520 |
seminiferous tubule development(GO:0072520) |
| 0.1 |
0.4 |
GO:0035964 |
COPI-coated vesicle budding(GO:0035964) |
| 0.1 |
0.9 |
GO:1901550 |
regulation of endothelial cell development(GO:1901550) regulation of establishment of endothelial barrier(GO:1903140) |
| 0.1 |
0.8 |
GO:0032049 |
cardiolipin biosynthetic process(GO:0032049) |
| 0.1 |
2.1 |
GO:0001886 |
endothelial cell morphogenesis(GO:0001886) |
| 0.1 |
1.8 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 |
0.7 |
GO:0033353 |
S-adenosylmethionine cycle(GO:0033353) |
| 0.1 |
2.6 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 |
1.3 |
GO:0019243 |
methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 |
6.5 |
GO:0080171 |
lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.1 |
0.3 |
GO:1904457 |
positive regulation of neuronal action potential(GO:1904457) |
| 0.1 |
2.3 |
GO:0045662 |
negative regulation of myoblast differentiation(GO:0045662) cellular response to fatty acid(GO:0071398) |
| 0.1 |
0.2 |
GO:0060743 |
estrous cycle(GO:0044849) epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 |
2.6 |
GO:0090083 |
regulation of inclusion body assembly(GO:0090083) |
| 0.1 |
5.5 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 |
0.5 |
GO:0035092 |
sperm chromatin condensation(GO:0035092) |
| 0.1 |
1.0 |
GO:0060746 |
parental behavior(GO:0060746) |
| 0.1 |
2.7 |
GO:0090162 |
establishment of epithelial cell polarity(GO:0090162) |
| 0.1 |
0.8 |
GO:0046688 |
response to copper ion(GO:0046688) |
| 0.1 |
1.5 |
GO:0032292 |
myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.1 |
0.6 |
GO:0006120 |
mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 |
0.5 |
GO:0048664 |
neuron fate determination(GO:0048664) |
| 0.0 |
0.1 |
GO:0015014 |
heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 |
1.2 |
GO:0006625 |
protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 |
0.6 |
GO:0000291 |
nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.0 |
0.2 |
GO:0044314 |
protein K27-linked ubiquitination(GO:0044314) |
| 0.0 |
1.0 |
GO:0071385 |
cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 |
0.7 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 |
0.4 |
GO:0045187 |
regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 |
0.1 |
GO:0072344 |
nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) rescue of stalled ribosome(GO:0072344) |
| 0.0 |
0.1 |
GO:0015886 |
heme transport(GO:0015886) |
| 0.0 |
0.9 |
GO:0050819 |
negative regulation of coagulation(GO:0050819) |
| 0.0 |
0.1 |
GO:0044828 |
negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 |
0.5 |
GO:0051127 |
positive regulation of actin nucleation(GO:0051127) |
| 0.0 |
0.5 |
GO:0016973 |
poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 |
0.5 |
GO:0042773 |
ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 |
1.0 |
GO:0019228 |
neuronal action potential(GO:0019228) |
| 0.0 |
0.2 |
GO:0045974 |
miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 |
0.4 |
GO:0007097 |
nuclear migration(GO:0007097) |
| 0.0 |
1.1 |
GO:0031338 |
regulation of vesicle fusion(GO:0031338) |
| 0.0 |
0.4 |
GO:0033194 |
response to hydroperoxide(GO:0033194) |
| 0.0 |
0.1 |
GO:0060018 |
astrocyte fate commitment(GO:0060018) |
| 0.0 |
0.7 |
GO:0006940 |
regulation of smooth muscle contraction(GO:0006940) |
| 0.0 |
0.6 |
GO:0006383 |
transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 |
0.4 |
GO:0000470 |
maturation of LSU-rRNA(GO:0000470) |
| 0.0 |
1.1 |
GO:0050852 |
T cell receptor signaling pathway(GO:0050852) |
| 0.0 |
1.0 |
GO:0048146 |
positive regulation of fibroblast proliferation(GO:0048146) |
| 0.0 |
0.3 |
GO:1902043 |
positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 |
0.2 |
GO:0022904 |
respiratory electron transport chain(GO:0022904) |
| 0.0 |
0.2 |
GO:0061469 |
regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 |
0.9 |
GO:0051965 |
positive regulation of synapse assembly(GO:0051965) |
| 0.0 |
0.2 |
GO:0030497 |
fatty acid elongation(GO:0030497) |
| 0.0 |
0.1 |
GO:0046686 |
response to cadmium ion(GO:0046686) |
| 0.0 |
0.4 |
GO:0034605 |
cellular response to heat(GO:0034605) |
| 0.0 |
1.2 |
GO:0098780 |
mitophagy in response to mitochondrial depolarization(GO:0098779) response to mitochondrial depolarisation(GO:0098780) |