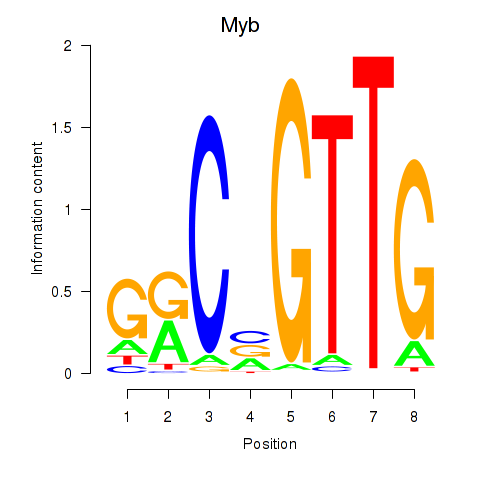
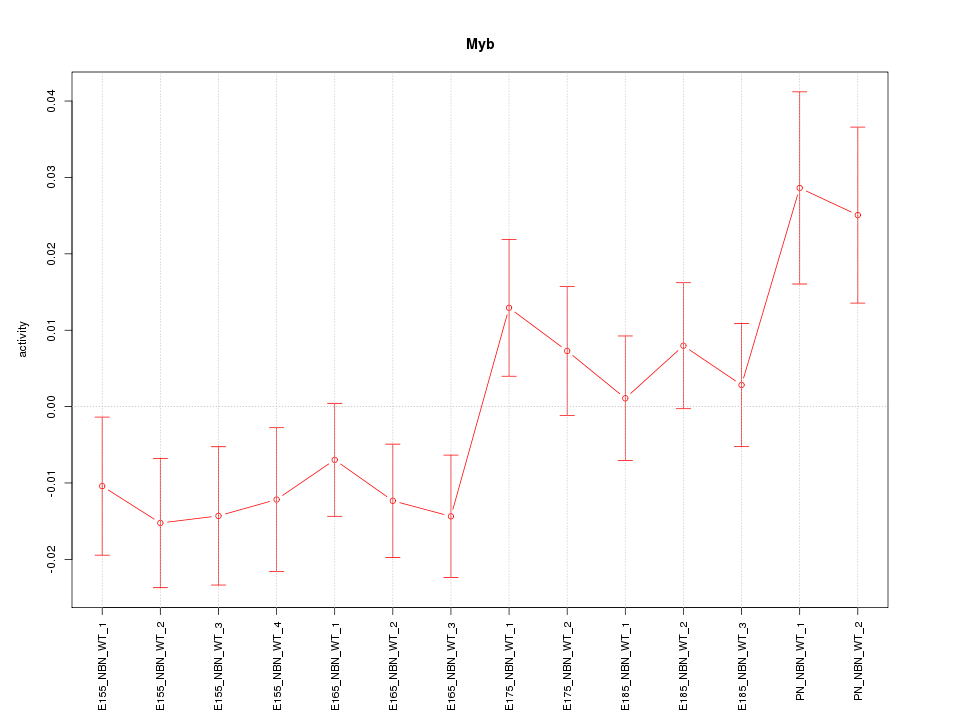
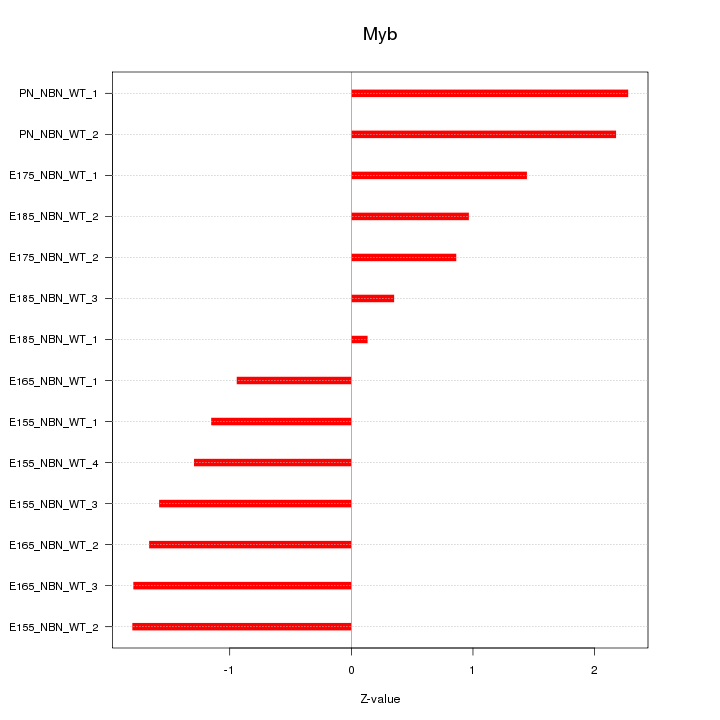
| 0.7 |
3.7 |
GO:0070460 |
thyroid-stimulating hormone secretion(GO:0070460) |
| 0.7 |
2.2 |
GO:0007341 |
penetration of zona pellucida(GO:0007341) |
| 0.7 |
4.4 |
GO:0035469 |
determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.6 |
2.3 |
GO:2000427 |
positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.5 |
2.0 |
GO:0072385 |
minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.4 |
2.6 |
GO:0033600 |
negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.4 |
4.2 |
GO:1901223 |
negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.4 |
2.0 |
GO:0047484 |
regulation of response to osmotic stress(GO:0047484) |
| 0.4 |
1.1 |
GO:1901896 |
positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.3 |
1.7 |
GO:0031915 |
positive regulation of synaptic plasticity(GO:0031915) |
| 0.3 |
2.1 |
GO:0019695 |
choline metabolic process(GO:0019695) |
| 0.3 |
0.9 |
GO:0034162 |
toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.3 |
0.9 |
GO:2000564 |
regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) CD8-positive, alpha-beta T cell proliferation(GO:0035740) positive regulation of interleukin-12 biosynthetic process(GO:0045084) negative regulation of regulatory T cell differentiation(GO:0045590) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.3 |
0.8 |
GO:1903903 |
regulation of establishment of T cell polarity(GO:1903903) |
| 0.3 |
2.3 |
GO:0048752 |
semicircular canal morphogenesis(GO:0048752) |
| 0.2 |
1.2 |
GO:0001923 |
B-1 B cell differentiation(GO:0001923) |
| 0.2 |
0.9 |
GO:0007056 |
spindle assembly involved in female meiosis(GO:0007056) positive regulation of oocyte development(GO:0060282) |
| 0.2 |
1.4 |
GO:0032483 |
regulation of Rab protein signal transduction(GO:0032483) |
| 0.2 |
1.3 |
GO:0034316 |
negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.2 |
0.6 |
GO:0006556 |
S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 |
1.1 |
GO:0006957 |
complement activation, alternative pathway(GO:0006957) |
| 0.2 |
1.4 |
GO:0035735 |
intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.2 |
1.1 |
GO:1900246 |
positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.2 |
0.9 |
GO:0045657 |
positive regulation of monocyte differentiation(GO:0045657) cellular response to potassium ion starvation(GO:0051365) |
| 0.2 |
0.5 |
GO:0045204 |
MAPK export from nucleus(GO:0045204) |
| 0.2 |
0.3 |
GO:2000766 |
negative regulation of cytoplasmic translation(GO:2000766) |
| 0.2 |
0.9 |
GO:0010032 |
meiotic chromosome condensation(GO:0010032) |
| 0.2 |
0.7 |
GO:0021914 |
negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.2 |
1.7 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 0.2 |
0.5 |
GO:1903244 |
positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.2 |
0.5 |
GO:0014028 |
notochord formation(GO:0014028) |
| 0.2 |
1.2 |
GO:0000395 |
mRNA 5'-splice site recognition(GO:0000395) |
| 0.2 |
2.4 |
GO:0022027 |
interkinetic nuclear migration(GO:0022027) astral microtubule organization(GO:0030953) |
| 0.1 |
1.0 |
GO:0019254 |
carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.1 |
3.1 |
GO:0001522 |
pseudouridine synthesis(GO:0001522) |
| 0.1 |
0.4 |
GO:1903225 |
negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 |
2.3 |
GO:0038166 |
angiotensin-activated signaling pathway(GO:0038166) |
| 0.1 |
0.8 |
GO:0006528 |
asparagine metabolic process(GO:0006528) |
| 0.1 |
0.4 |
GO:0061153 |
trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.1 |
2.1 |
GO:0070986 |
left/right axis specification(GO:0070986) |
| 0.1 |
0.7 |
GO:1903361 |
protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 |
0.9 |
GO:0006297 |
nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 |
1.2 |
GO:0046036 |
GTP biosynthetic process(GO:0006183) CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.1 |
0.8 |
GO:0036159 |
inner dynein arm assembly(GO:0036159) |
| 0.1 |
0.5 |
GO:0051725 |
protein de-ADP-ribosylation(GO:0051725) |
| 0.1 |
1.4 |
GO:0019985 |
translesion synthesis(GO:0019985) |
| 0.1 |
0.3 |
GO:0035621 |
ER to Golgi ceramide transport(GO:0035621) ceramide transport(GO:0035627) |
| 0.1 |
0.1 |
GO:0060854 |
patterning of lymph vessels(GO:0060854) |
| 0.1 |
1.6 |
GO:0021891 |
olfactory bulb interneuron development(GO:0021891) |
| 0.1 |
0.5 |
GO:0014022 |
neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.1 |
0.3 |
GO:0001866 |
NK T cell proliferation(GO:0001866) |
| 0.1 |
9.1 |
GO:0048663 |
neuron fate commitment(GO:0048663) |
| 0.1 |
1.0 |
GO:0002643 |
regulation of tolerance induction(GO:0002643) |
| 0.1 |
0.4 |
GO:0030997 |
regulation of centriole-centriole cohesion(GO:0030997) |
| 0.1 |
0.5 |
GO:0051189 |
Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.1 |
0.9 |
GO:0010457 |
centriole-centriole cohesion(GO:0010457) |
| 0.1 |
0.7 |
GO:0043983 |
histone H4-K12 acetylation(GO:0043983) |
| 0.1 |
0.5 |
GO:0032815 |
negative regulation of natural killer cell activation(GO:0032815) |
| 0.1 |
0.6 |
GO:0010571 |
positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 |
1.2 |
GO:0051988 |
regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.1 |
0.2 |
GO:0070172 |
epithelial-mesenchymal cell signaling(GO:0060684) positive regulation of tooth mineralization(GO:0070172) |
| 0.1 |
0.4 |
GO:0070966 |
nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 |
1.3 |
GO:0032515 |
negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 |
0.3 |
GO:0006624 |
vacuolar protein processing(GO:0006624) |
| 0.1 |
0.7 |
GO:0097034 |
mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 |
1.2 |
GO:0010603 |
regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.1 |
0.9 |
GO:0015937 |
coenzyme A biosynthetic process(GO:0015937) |
| 0.1 |
1.5 |
GO:0010172 |
embryonic body morphogenesis(GO:0010172) |
| 0.1 |
0.9 |
GO:0007063 |
regulation of sister chromatid cohesion(GO:0007063) |
| 0.1 |
1.7 |
GO:0034724 |
DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.1 |
0.3 |
GO:0060040 |
retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 |
0.3 |
GO:0071896 |
protein localization to adherens junction(GO:0071896) |
| 0.1 |
1.1 |
GO:0090161 |
Golgi ribbon formation(GO:0090161) |
| 0.1 |
0.3 |
GO:0016576 |
histone dephosphorylation(GO:0016576) |
| 0.1 |
1.0 |
GO:0042249 |
establishment of planar polarity of embryonic epithelium(GO:0042249) |
| 0.1 |
0.3 |
GO:0070166 |
enamel mineralization(GO:0070166) |
| 0.1 |
2.1 |
GO:0048255 |
mRNA stabilization(GO:0048255) |
| 0.1 |
0.1 |
GO:1903026 |
negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.1 |
1.7 |
GO:0007638 |
mechanosensory behavior(GO:0007638) |
| 0.1 |
0.5 |
GO:0000712 |
resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 |
0.2 |
GO:2001170 |
negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.0 |
0.7 |
GO:0009072 |
aromatic amino acid family metabolic process(GO:0009072) |
| 0.0 |
0.6 |
GO:0034501 |
protein localization to kinetochore(GO:0034501) |
| 0.0 |
0.3 |
GO:0034587 |
piRNA metabolic process(GO:0034587) |
| 0.0 |
0.8 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 |
0.2 |
GO:1902965 |
regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.0 |
0.4 |
GO:0035520 |
monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 |
0.3 |
GO:0043982 |
histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 |
0.5 |
GO:2000780 |
negative regulation of double-strand break repair(GO:2000780) |
| 0.0 |
0.3 |
GO:0000467 |
exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.0 |
0.1 |
GO:0061086 |
negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 |
0.8 |
GO:2000060 |
positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.0 |
0.6 |
GO:0045793 |
positive regulation of cell size(GO:0045793) |
| 0.0 |
0.7 |
GO:0034508 |
centromere complex assembly(GO:0034508) |
| 0.0 |
0.3 |
GO:0036120 |
cellular response to platelet-derived growth factor stimulus(GO:0036120) positive regulation of tyrosine phosphorylation of Stat5 protein(GO:0042523) |
| 0.0 |
0.3 |
GO:0033299 |
secretion of lysosomal enzymes(GO:0033299) |
| 0.0 |
0.4 |
GO:0060049 |
regulation of protein glycosylation(GO:0060049) |
| 0.0 |
0.4 |
GO:0034497 |
protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 |
0.5 |
GO:0030252 |
growth hormone secretion(GO:0030252) |
| 0.0 |
0.2 |
GO:0006398 |
mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 |
0.8 |
GO:0090162 |
establishment of epithelial cell polarity(GO:0090162) |
| 0.0 |
0.5 |
GO:0002021 |
response to dietary excess(GO:0002021) |
| 0.0 |
1.1 |
GO:0030514 |
negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 |
6.3 |
GO:0051480 |
regulation of cytosolic calcium ion concentration(GO:0051480) |
| 0.0 |
0.4 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 |
0.2 |
GO:0015868 |
purine ribonucleotide transport(GO:0015868) |
| 0.0 |
1.5 |
GO:0045669 |
positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 |
0.2 |
GO:0046085 |
adenosine metabolic process(GO:0046085) |
| 0.0 |
0.0 |
GO:0030214 |
hyaluronan catabolic process(GO:0030214) |
| 0.0 |
0.2 |
GO:0071340 |
skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 |
0.3 |
GO:0019243 |
methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 |
0.0 |
GO:0060830 |
ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 |
0.5 |
GO:0022617 |
extracellular matrix disassembly(GO:0022617) |
| 0.0 |
0.5 |
GO:1902236 |
negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 |
0.2 |
GO:1990126 |
retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 |
0.3 |
GO:0006465 |
signal peptide processing(GO:0006465) |
| 0.0 |
0.1 |
GO:0007021 |
tubulin complex assembly(GO:0007021) |
| 0.0 |
0.3 |
GO:0032981 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 |
1.1 |
GO:0007127 |
meiosis I(GO:0007127) |
| 0.0 |
0.3 |
GO:0044381 |
glucose import in response to insulin stimulus(GO:0044381) regulation of glucose import in response to insulin stimulus(GO:2001273) |
| 0.0 |
0.5 |
GO:0031122 |
cytoplasmic microtubule organization(GO:0031122) |
| 0.0 |
0.2 |
GO:0046686 |
response to cadmium ion(GO:0046686) |
| 0.0 |
0.3 |
GO:0032467 |
positive regulation of cytokinesis(GO:0032467) |
| 0.0 |
0.2 |
GO:0071392 |
cellular response to estradiol stimulus(GO:0071392) |
| 0.0 |
0.0 |
GO:0018076 |
N-terminal peptidyl-lysine acetylation(GO:0018076) positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 |
0.3 |
GO:0006611 |
protein export from nucleus(GO:0006611) |
| 0.0 |
0.4 |
GO:0008584 |
male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.0 |
0.2 |
GO:0048026 |
positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 |
0.4 |
GO:0000381 |
regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 |
0.6 |
GO:0046324 |
regulation of glucose import(GO:0046324) |
| 0.0 |
1.0 |
GO:0030010 |
establishment of cell polarity(GO:0030010) |
| 0.0 |
0.5 |
GO:0006749 |
glutathione metabolic process(GO:0006749) |
| 0.0 |
1.1 |
GO:0007050 |
cell cycle arrest(GO:0007050) |
| 0.0 |
0.1 |
GO:0035887 |
aortic smooth muscle cell differentiation(GO:0035887) |