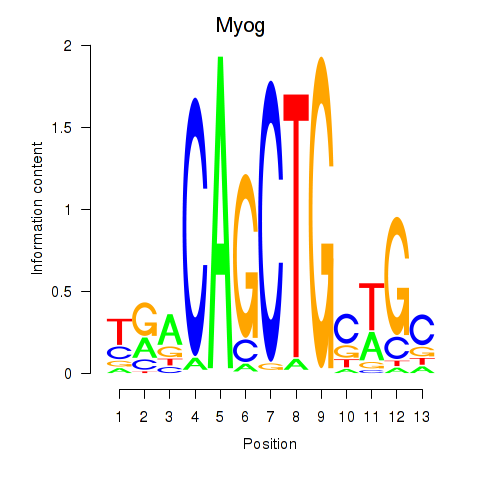
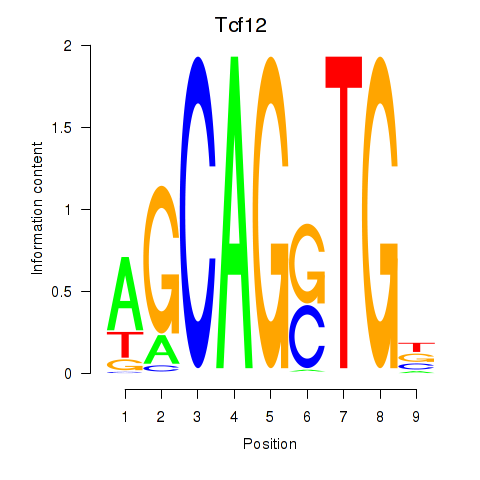
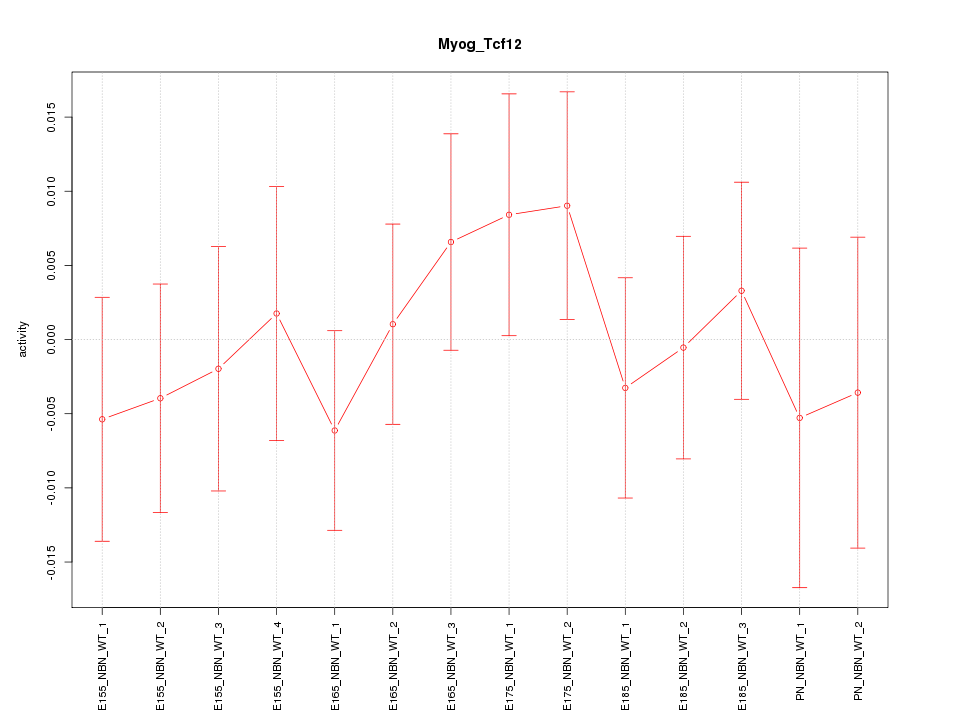
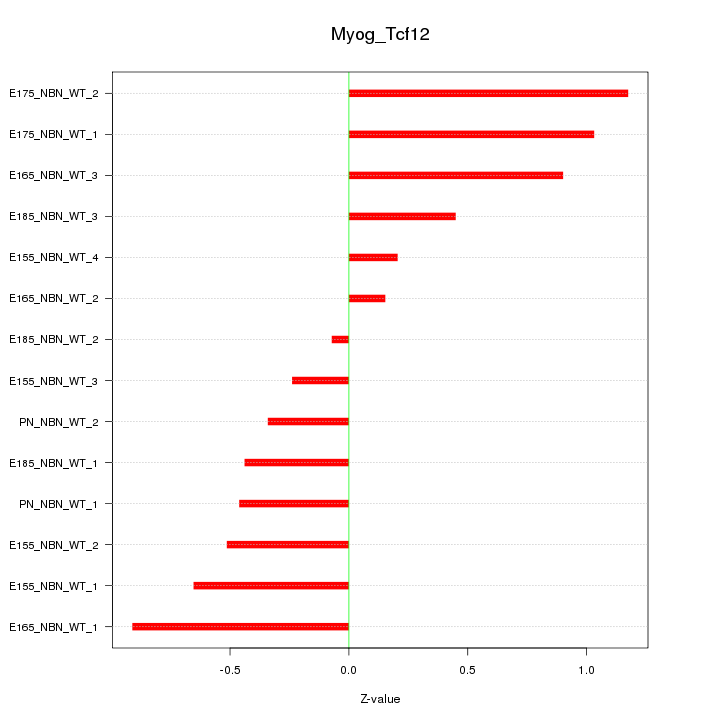
| 0.4 |
1.1 |
GO:0003345 |
proepicardium cell migration involved in pericardium morphogenesis(GO:0003345) |
| 0.3 |
2.9 |
GO:0036371 |
protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.3 |
1.7 |
GO:0021773 |
striatal medium spiny neuron differentiation(GO:0021773) |
| 0.3 |
0.8 |
GO:0006601 |
creatine biosynthetic process(GO:0006601) |
| 0.3 |
0.6 |
GO:0045110 |
intermediate filament bundle assembly(GO:0045110) |
| 0.2 |
0.7 |
GO:0071374 |
cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.2 |
1.4 |
GO:0050861 |
positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.2 |
1.0 |
GO:0086013 |
membrane repolarization during cardiac muscle cell action potential(GO:0086013) |
| 0.2 |
0.8 |
GO:0014012 |
peripheral nervous system axon regeneration(GO:0014012) |
| 0.2 |
0.6 |
GO:1903048 |
regulation of acetylcholine-gated cation channel activity(GO:1903048) |
| 0.2 |
0.8 |
GO:0032423 |
regulation of mismatch repair(GO:0032423) |
| 0.1 |
0.4 |
GO:0050925 |
negative regulation of negative chemotaxis(GO:0050925) |
| 0.1 |
0.7 |
GO:1901979 |
negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.1 |
0.2 |
GO:0007158 |
neuron cell-cell adhesion(GO:0007158) |
| 0.1 |
0.4 |
GO:1903587 |
regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.1 |
0.4 |
GO:0016340 |
calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.1 |
0.3 |
GO:0032430 |
positive regulation of phospholipase A2 activity(GO:0032430) activation of meiosis involved in egg activation(GO:0060466) |
| 0.1 |
0.3 |
GO:0048686 |
regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.1 |
0.5 |
GO:0001778 |
plasma membrane repair(GO:0001778) |
| 0.1 |
0.3 |
GO:0021577 |
hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.1 |
0.3 |
GO:0021586 |
pons maturation(GO:0021586) |
| 0.1 |
0.3 |
GO:0045113 |
regulation of integrin biosynthetic process(GO:0045113) |
| 0.1 |
0.2 |
GO:0061153 |
trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.1 |
0.4 |
GO:2000809 |
positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.1 |
0.4 |
GO:0048133 |
germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 |
0.2 |
GO:1904049 |
regulation of spontaneous neurotransmitter secretion(GO:1904048) negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.1 |
0.2 |
GO:0019085 |
early viral transcription(GO:0019085) |
| 0.1 |
0.2 |
GO:0001830 |
trophectodermal cell fate commitment(GO:0001830) |
| 0.1 |
0.2 |
GO:0018199 |
peptidyl-glutamine modification(GO:0018199) |
| 0.1 |
0.3 |
GO:0090038 |
negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 |
0.5 |
GO:1900042 |
positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 |
0.2 |
GO:1903334 |
positive regulation of protein folding(GO:1903334) |
| 0.1 |
0.2 |
GO:0046959 |
habituation(GO:0046959) |
| 0.1 |
1.4 |
GO:2000310 |
regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.1 |
1.0 |
GO:0035235 |
ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 |
0.2 |
GO:0044339 |
canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.1 |
0.2 |
GO:1903244 |
positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.1 |
0.2 |
GO:0009174 |
UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.1 |
0.2 |
GO:0019405 |
alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 |
0.2 |
GO:2000507 |
positive regulation of energy homeostasis(GO:2000507) |
| 0.1 |
0.2 |
GO:0051305 |
chromosome movement towards spindle pole(GO:0051305) |
| 0.1 |
0.3 |
GO:0060591 |
chondroblast differentiation(GO:0060591) |
| 0.1 |
0.2 |
GO:0000415 |
negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 |
0.2 |
GO:0098528 |
skeletal muscle fiber differentiation(GO:0098528) |
| 0.1 |
0.1 |
GO:0060025 |
regulation of synaptic activity(GO:0060025) |
| 0.0 |
0.3 |
GO:0032534 |
regulation of microvillus assembly(GO:0032534) |
| 0.0 |
0.4 |
GO:0031547 |
brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 |
0.0 |
GO:1900451 |
positive regulation of glutamate receptor signaling pathway(GO:1900451) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.0 |
0.2 |
GO:0090168 |
Golgi reassembly(GO:0090168) |
| 0.0 |
0.3 |
GO:0007342 |
fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 |
0.2 |
GO:1900122 |
positive regulation of receptor binding(GO:1900122) |
| 0.0 |
0.1 |
GO:0015882 |
L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.0 |
0.2 |
GO:0045188 |
regulation of circadian sleep/wake cycle, non-REM sleep(GO:0045188) |
| 0.0 |
0.4 |
GO:0018095 |
protein polyglutamylation(GO:0018095) |
| 0.0 |
0.9 |
GO:0010614 |
negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 |
0.2 |
GO:0010724 |
regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 |
1.4 |
GO:0061003 |
positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 |
0.2 |
GO:0045218 |
zonula adherens maintenance(GO:0045218) |
| 0.0 |
0.1 |
GO:0051542 |
elastin biosynthetic process(GO:0051542) |
| 0.0 |
0.0 |
GO:0060423 |
foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.0 |
0.3 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.0 |
0.7 |
GO:0018149 |
peptide cross-linking(GO:0018149) |
| 0.0 |
0.0 |
GO:0090238 |
positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.0 |
0.4 |
GO:0033631 |
cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 |
0.3 |
GO:0060923 |
cardiac muscle cell fate commitment(GO:0060923) |
| 0.0 |
0.0 |
GO:0098911 |
regulation of ventricular cardiac muscle cell action potential(GO:0098911) |
| 0.0 |
0.1 |
GO:2000286 |
receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 |
1.0 |
GO:0031571 |
mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 |
0.1 |
GO:0060715 |
syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.0 |
0.1 |
GO:0033615 |
mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 |
0.1 |
GO:0009193 |
pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.0 |
0.2 |
GO:0016080 |
synaptic vesicle targeting(GO:0016080) |
| 0.0 |
0.5 |
GO:0045161 |
neuronal ion channel clustering(GO:0045161) |
| 0.0 |
0.2 |
GO:0016081 |
synaptic vesicle docking(GO:0016081) |
| 0.0 |
0.5 |
GO:0051127 |
positive regulation of actin nucleation(GO:0051127) |
| 0.0 |
0.2 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.0 |
0.3 |
GO:1901844 |
regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 |
0.2 |
GO:1902414 |
protein localization to cell junction(GO:1902414) |
| 0.0 |
0.2 |
GO:0032224 |
positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.0 |
0.1 |
GO:0097089 |
methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.0 |
0.2 |
GO:0003419 |
growth plate cartilage chondrocyte proliferation(GO:0003419) |
| 0.0 |
0.3 |
GO:0018230 |
peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 |
0.2 |
GO:0000429 |
carbon catabolite regulation of transcription from RNA polymerase II promoter(GO:0000429) carbon catabolite activation of transcription from RNA polymerase II promoter(GO:0000436) |
| 0.0 |
0.1 |
GO:0019287 |
isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.0 |
0.2 |
GO:0097151 |
positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 |
0.1 |
GO:0060912 |
cardiac cell fate specification(GO:0060912) |
| 0.0 |
0.1 |
GO:0015014 |
heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 |
0.1 |
GO:0010459 |
negative regulation of heart rate(GO:0010459) |
| 0.0 |
0.3 |
GO:0006706 |
steroid catabolic process(GO:0006706) |
| 0.0 |
0.0 |
GO:1901420 |
negative regulation of response to alcohol(GO:1901420) |
| 0.0 |
0.1 |
GO:1904690 |
regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.0 |
0.1 |
GO:0019805 |
quinolinate biosynthetic process(GO:0019805) |
| 0.0 |
0.1 |
GO:0060017 |
parathyroid gland development(GO:0060017) |
| 0.0 |
0.1 |
GO:1902237 |
positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902237) |
| 0.0 |
0.1 |
GO:0046898 |
response to cycloheximide(GO:0046898) |
| 0.0 |
0.1 |
GO:0060596 |
mammary placode formation(GO:0060596) |
| 0.0 |
0.1 |
GO:0007181 |
transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 |
0.0 |
GO:0032227 |
negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 |
0.1 |
GO:0035622 |
intrahepatic bile duct development(GO:0035622) positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.0 |
0.1 |
GO:0001927 |
exocyst assembly(GO:0001927) |
| 0.0 |
0.1 |
GO:0072718 |
response to cisplatin(GO:0072718) |
| 0.0 |
0.3 |
GO:0045898 |
regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 |
0.1 |
GO:0060399 |
positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 |
0.2 |
GO:0045056 |
transcytosis(GO:0045056) |
| 0.0 |
2.0 |
GO:0030032 |
lamellipodium assembly(GO:0030032) |
| 0.0 |
0.1 |
GO:0035331 |
negative regulation of hippo signaling(GO:0035331) |
| 0.0 |
0.2 |
GO:0007200 |
phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 |
0.2 |
GO:0006620 |
posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 |
0.0 |
GO:0002331 |
pre-B cell allelic exclusion(GO:0002331) |
| 0.0 |
0.3 |
GO:0048172 |
regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 |
0.0 |
GO:1902474 |
positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 |
0.1 |
GO:0010499 |
proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 |
0.0 |
GO:0038095 |
Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.0 |
0.5 |
GO:0008210 |
estrogen metabolic process(GO:0008210) |
| 0.0 |
0.1 |
GO:0035574 |
histone H4-K20 demethylation(GO:0035574) |
| 0.0 |
0.4 |
GO:0032897 |
negative regulation of viral transcription(GO:0032897) |
| 0.0 |
0.1 |
GO:0070305 |
response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) |
| 0.0 |
0.1 |
GO:0032483 |
regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 |
0.2 |
GO:2000465 |
regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.0 |
0.5 |
GO:0001573 |
ganglioside metabolic process(GO:0001573) |
| 0.0 |
0.2 |
GO:0014832 |
urinary bladder smooth muscle contraction(GO:0014832) |
| 0.0 |
0.1 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 |
0.2 |
GO:1904153 |
negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 |
0.1 |
GO:0002002 |
regulation of angiotensin levels in blood(GO:0002002) |
| 0.0 |
0.1 |
GO:1903288 |
regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.0 |
0.0 |
GO:0035509 |
negative regulation of myosin-light-chain-phosphatase activity(GO:0035509) |
| 0.0 |
0.1 |
GO:0046826 |
negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 |
0.2 |
GO:0030206 |
chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 |
0.2 |
GO:0006883 |
cellular sodium ion homeostasis(GO:0006883) |
| 0.0 |
0.1 |
GO:0032485 |
regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 |
0.1 |
GO:0070966 |
nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 |
0.1 |
GO:0033326 |
cerebrospinal fluid secretion(GO:0033326) |
| 0.0 |
0.1 |
GO:0035735 |
intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 |
0.3 |
GO:0035563 |
positive regulation of chromatin binding(GO:0035563) |
| 0.0 |
0.5 |
GO:0050873 |
brown fat cell differentiation(GO:0050873) |
| 0.0 |
0.1 |
GO:0034140 |
negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) aggrephagy(GO:0035973) |
| 0.0 |
0.2 |
GO:2000251 |
positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 |
0.2 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.0 |
0.1 |
GO:0006295 |
nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 |
0.1 |
GO:0034587 |
piRNA metabolic process(GO:0034587) |
| 0.0 |
0.1 |
GO:0060743 |
epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 |
0.1 |
GO:1901660 |
calcium ion export(GO:1901660) |
| 0.0 |
0.0 |
GO:0044333 |
Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.0 |
0.1 |
GO:0036506 |
maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 |
0.1 |
GO:0010668 |
ectodermal cell differentiation(GO:0010668) |
| 0.0 |
0.2 |
GO:0003157 |
endocardium development(GO:0003157) |
| 0.0 |
0.7 |
GO:0019228 |
neuronal action potential(GO:0019228) |
| 0.0 |
0.1 |
GO:0045347 |
negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 |
0.1 |
GO:0000480 |
endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 |
0.1 |
GO:0045059 |
positive thymic T cell selection(GO:0045059) |
| 0.0 |
0.2 |
GO:0021527 |
spinal cord association neuron differentiation(GO:0021527) |
| 0.0 |
0.1 |
GO:0071787 |
endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 |
0.0 |
GO:2000393 |
negative regulation of lamellipodium organization(GO:1902744) negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.0 |
0.2 |
GO:0097119 |
postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 |
0.2 |
GO:0021785 |
branchiomotor neuron axon guidance(GO:0021785) |
| 0.0 |
0.1 |
GO:0009650 |
UV protection(GO:0009650) |
| 0.0 |
0.1 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 |
0.1 |
GO:0060263 |
regulation of respiratory burst(GO:0060263) positive regulation of respiratory burst(GO:0060267) |
| 0.0 |
0.1 |
GO:2000786 |
positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 |
0.1 |
GO:1904008 |
response to monosodium glutamate(GO:1904008) cellular response to monosodium glutamate(GO:1904009) |
| 0.0 |
0.4 |
GO:0007616 |
long-term memory(GO:0007616) |
| 0.0 |
0.2 |
GO:0030157 |
pancreatic juice secretion(GO:0030157) |
| 0.0 |
0.3 |
GO:0031116 |
positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 |
0.0 |
GO:1903998 |
regulation of eating behavior(GO:1903998) |
| 0.0 |
0.2 |
GO:0002495 |
antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 |
0.2 |
GO:0021819 |
layer formation in cerebral cortex(GO:0021819) |
| 0.0 |
0.2 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 |
0.0 |
GO:0035435 |
phosphate ion transmembrane transport(GO:0035435) |
| 0.0 |
0.1 |
GO:0010040 |
response to iron(II) ion(GO:0010040) |
| 0.0 |
0.1 |
GO:0033580 |
protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.0 |
0.1 |
GO:0050910 |
detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 |
0.2 |
GO:0080182 |
histone H3-K4 trimethylation(GO:0080182) |
| 0.0 |
0.2 |
GO:0006895 |
Golgi to endosome transport(GO:0006895) |
| 0.0 |
0.1 |
GO:1902047 |
polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) |
| 0.0 |
0.2 |
GO:0006415 |
translational termination(GO:0006415) |
| 0.0 |
0.0 |
GO:0070634 |
transepithelial ammonium transport(GO:0070634) |
| 0.0 |
0.2 |
GO:0051988 |
regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 |
0.0 |
GO:1902260 |
negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 |
0.1 |
GO:0007258 |
JUN phosphorylation(GO:0007258) |
| 0.0 |
0.1 |
GO:0070493 |
thrombin receptor signaling pathway(GO:0070493) |
| 0.0 |
0.1 |
GO:0015992 |
proton transport(GO:0015992) |
| 0.0 |
0.1 |
GO:0007202 |
activation of phospholipase C activity(GO:0007202) |
| 0.0 |
0.0 |
GO:0031581 |
hemidesmosome assembly(GO:0031581) |
| 0.0 |
0.1 |
GO:0035887 |
aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 |
0.1 |
GO:1901223 |
negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 |
0.0 |
GO:0060741 |
prostate gland stromal morphogenesis(GO:0060741) |
| 0.0 |
0.1 |
GO:0038031 |
non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 |
0.1 |
GO:0030539 |
male genitalia development(GO:0030539) plasminogen activation(GO:0031639) |
| 0.0 |
0.0 |
GO:0010847 |
regulation of chromatin assembly(GO:0010847) |
| 0.0 |
0.1 |
GO:0031115 |
negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 |
0.1 |
GO:0043589 |
skin morphogenesis(GO:0043589) |
| 0.0 |
0.1 |
GO:0036297 |
interstrand cross-link repair(GO:0036297) |
| 0.0 |
0.1 |
GO:0051409 |
response to nitrosative stress(GO:0051409) |
| 0.0 |
0.1 |
GO:0045176 |
apical protein localization(GO:0045176) |
| 0.0 |
0.4 |
GO:0007520 |
myoblast fusion(GO:0007520) |
| 0.0 |
0.0 |
GO:1902804 |
negative regulation of synaptic vesicle transport(GO:1902804) |
| 0.0 |
0.2 |
GO:0051382 |
kinetochore assembly(GO:0051382) |
| 0.0 |
0.3 |
GO:0034723 |
DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 |
0.1 |
GO:0051571 |
positive regulation of histone H3-K4 methylation(GO:0051571) |
| 0.0 |
0.1 |
GO:0014807 |
regulation of somitogenesis(GO:0014807) |
| 0.0 |
0.2 |
GO:1903861 |
regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.0 |
0.1 |
GO:0043388 |
positive regulation of DNA binding(GO:0043388) |