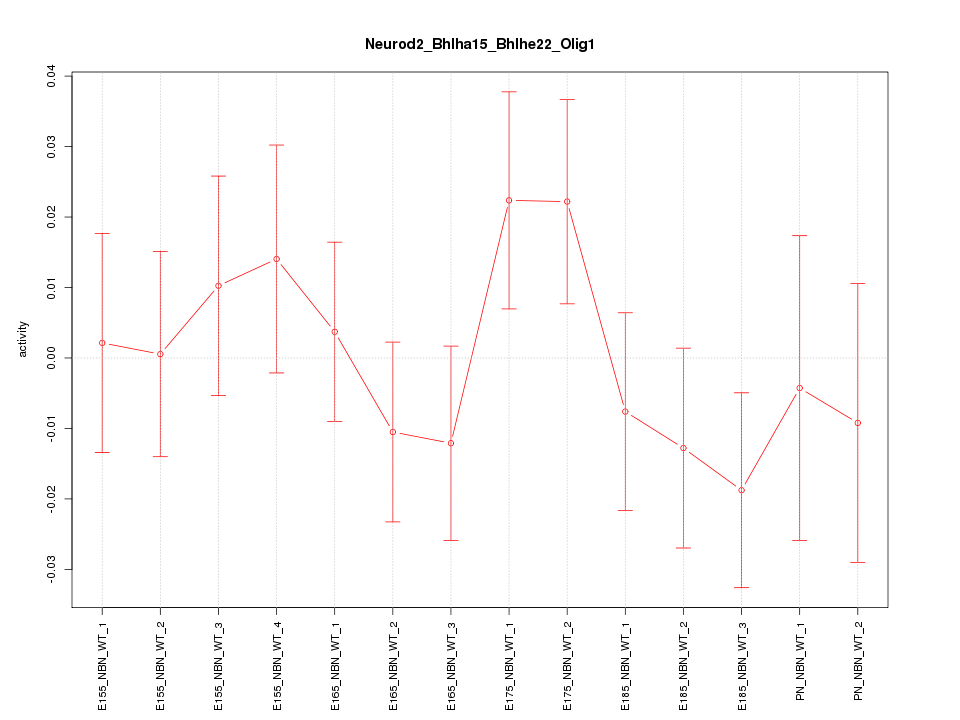
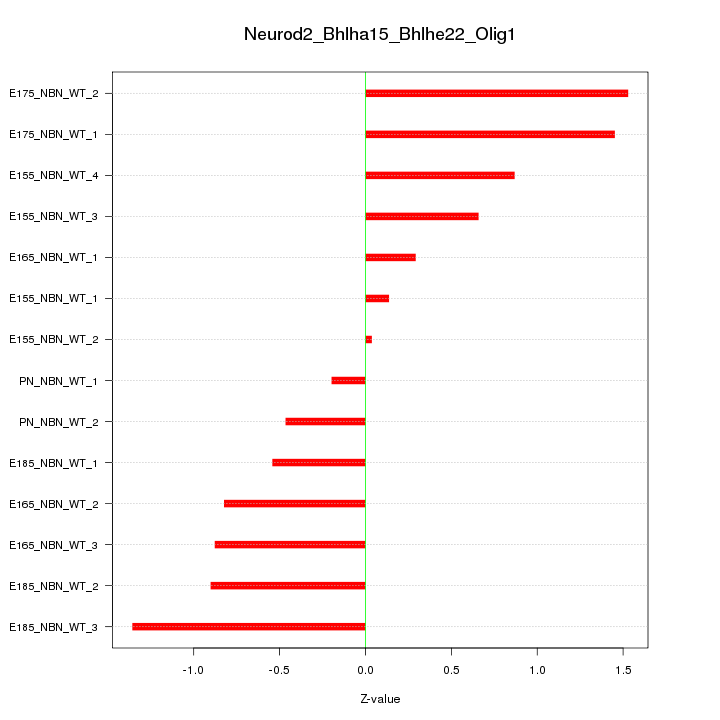
Motif ID: Neurod2_Bhlha15_Bhlhe22_Olig1
Z-value: 0.862
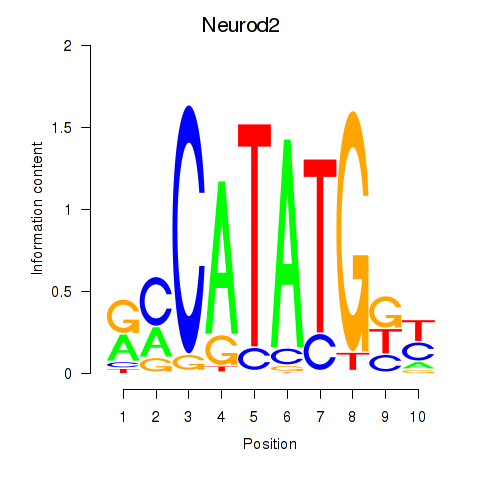
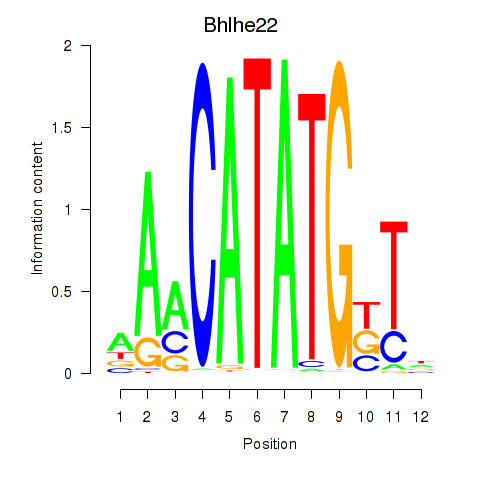
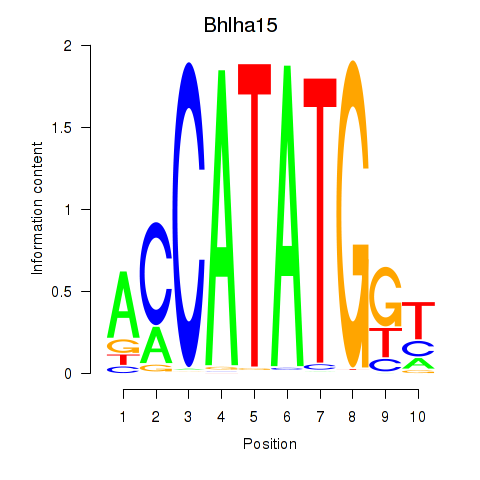
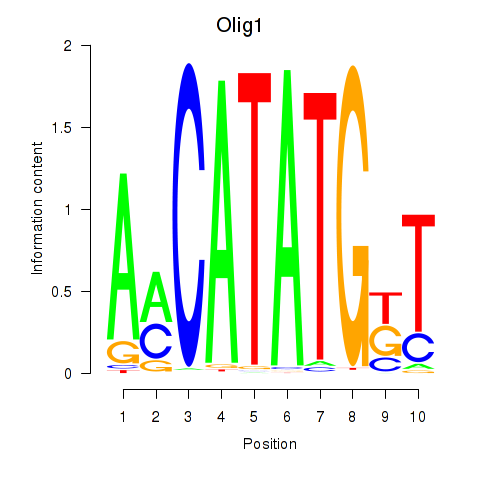
Transcription factors associated with Neurod2_Bhlha15_Bhlhe22_Olig1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Bhlha15 | ENSMUSG00000052271.7 | Bhlha15 |
| Bhlhe22 | ENSMUSG00000025128.6 | Bhlhe22 |
| Neurod2 | ENSMUSG00000038255.6 | Neurod2 |
| Olig1 | ENSMUSG00000046160.5 | Olig1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Neurod2 | mm10_v2_chr11_-_98329641_98329654 | 0.40 | 1.6e-01 | Click! |
| Bhlhe22 | mm10_v2_chr3_+_18054258_18054325 | 0.35 | 2.2e-01 | Click! |
| Olig1 | mm10_v2_chr16_+_91269759_91269778 | -0.21 | 4.8e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.2 | 0.7 | GO:0002159 | desmosome assembly(GO:0002159) adherens junction maintenance(GO:0034334) intermediate filament bundle assembly(GO:0045110) maintenance of organ identity(GO:0048496) |
| 0.2 | 0.5 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.2 | 0.3 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) |
| 0.2 | 1.5 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.4 | GO:0010911 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) |
| 0.1 | 0.4 | GO:0032240 | RNA import into nucleus(GO:0006404) mRNA export from nucleus in response to heat stress(GO:0031990) negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.1 | 0.6 | GO:2000302 | regulation of synaptic vesicle endocytosis(GO:1900242) positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.1 | 0.3 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.1 | 2.0 | GO:0045956 | positive regulation of vesicle fusion(GO:0031340) positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.1 | 1.0 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.1 | 0.3 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.1 | 1.5 | GO:0034390 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.1 | 0.4 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.1 | 1.9 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.1 | 0.8 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 0.7 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.2 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.0 | 0.4 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.0 | 0.3 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 0.3 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 | 0.0 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 | 0.3 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.5 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.0 | 0.3 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.5 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.3 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.2 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.0 | 2.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.7 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.2 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.0 | 0.2 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.6 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.3 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 0.2 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.4 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.6 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.0 | 0.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 2.0 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 0.1 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.0 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.4 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 1.1 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.2 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.1 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.1 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.0 | 0.2 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.0 | 0.0 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 1.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.4 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.1 | 1.7 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.4 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 0.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.9 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.5 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 2.0 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.2 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.1 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 1.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.2 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.8 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.1 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.2 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.0 | GO:0071942 | XPC complex(GO:0071942) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.8 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.2 | 1.5 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 0.4 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 0.3 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.1 | 0.9 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 0.7 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 1.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 2.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 2.0 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.3 | GO:0045703 | pinocarveol dehydrogenase activity(GO:0018446) chloral hydrate dehydrogenase activity(GO:0018447) hydroxymethylmethylsilanediol oxidase activity(GO:0018448) 1-phenylethanol dehydrogenase activity(GO:0018449) myrtenol dehydrogenase activity(GO:0018450) cis-1,2-dihydroxy-1,2-dihydro-8-carboxynaphthalene dehydrogenase activity(GO:0034522) 3-hydroxy-4-methyloctanoyl-CoA dehydrogenase activity(GO:0034582) 2-hydroxy-4-isopropenylcyclohexane-1-carboxyl-CoA dehydrogenase activity(GO:0034778) cis-9,10-dihydroanthracene-9,10-diol dehydrogenase activity(GO:0034817) citronellol dehydrogenase activity(GO:0034821) naphthyl-2-hydroxymethyl-succinyl-CoA dehydrogenase activity(GO:0034847) 2,4,4-trimethyl-1-pentanol dehydrogenase activity(GO:0034863) 2,4,4-trimethyl-3-hydroxypentanoyl-CoA dehydrogenase activity(GO:0034868) 1-hydroxy-4,4-dimethylpentan-3-one dehydrogenase activity(GO:0034871) endosulfan diol dehydrogenase activity(GO:0034891) endosulfan hydroxyether dehydrogenase activity(GO:0034901) 3-hydroxy-2-methylhexanoyl-CoA dehydrogenase activity(GO:0034918) 3-hydroxy-2,6-dimethyl-5-methylene-heptanoyl-CoA dehydrogenase activity(GO:0034944) versicolorin reductase activity(GO:0042469) ketoreductase activity(GO:0045703) |
| 0.0 | 0.2 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.4 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 1.1 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.5 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.0 | 0.4 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.3 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.3 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.3 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.4 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.3 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.1 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |