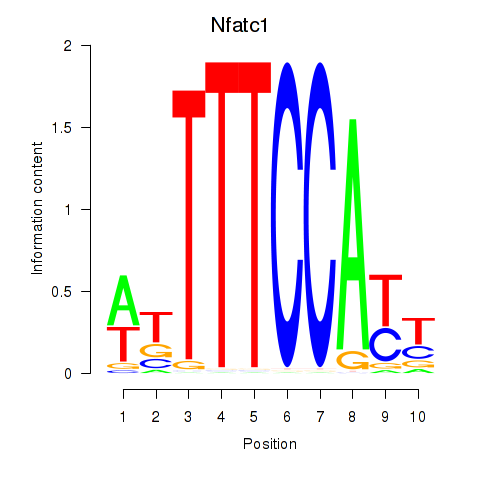
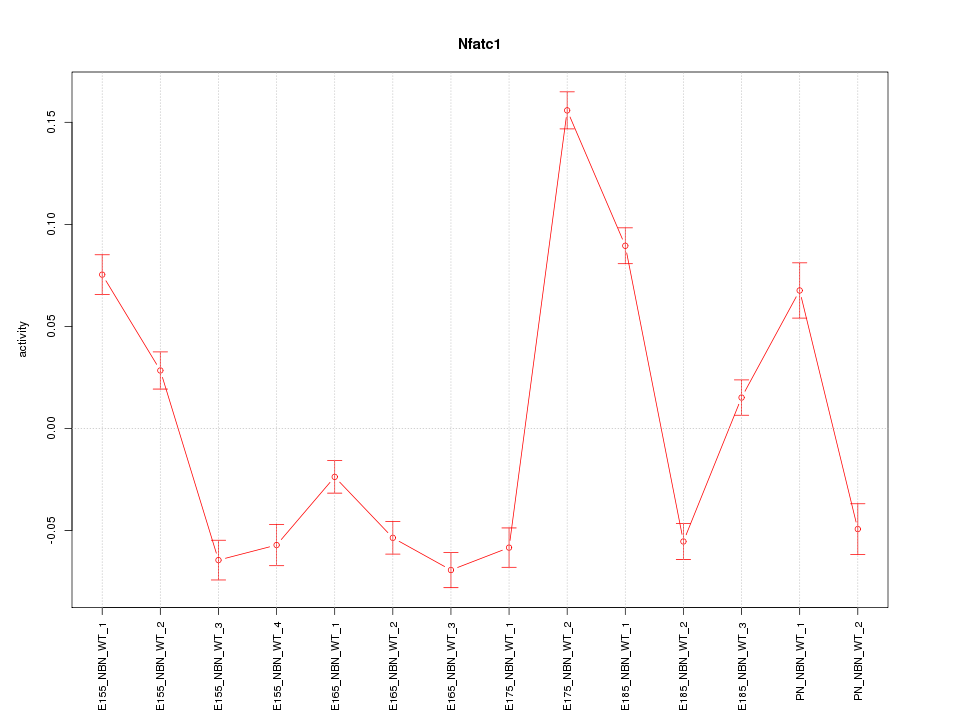
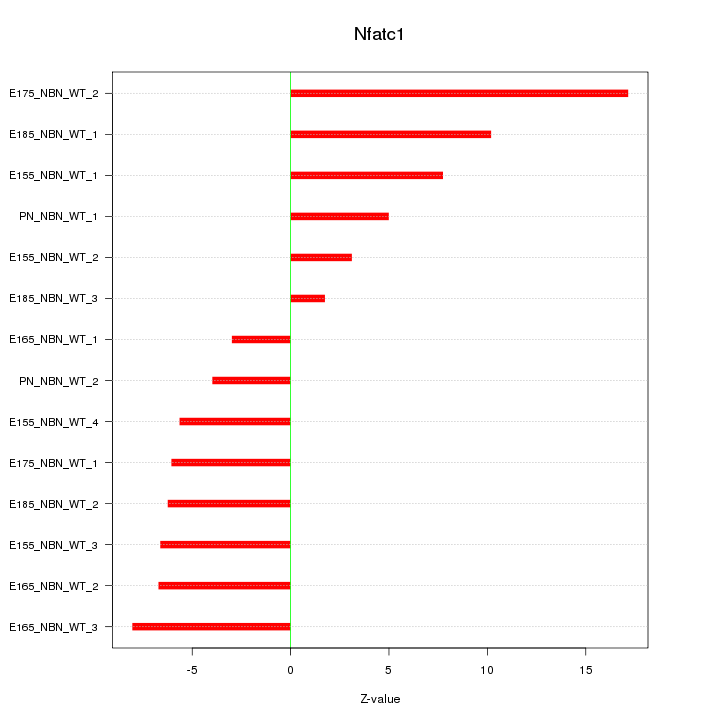
| 2.1 |
6.3 |
GO:1900158 |
negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 1.2 |
5.9 |
GO:0070327 |
thyroid hormone transport(GO:0070327) |
| 1.0 |
4.8 |
GO:0032224 |
positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.8 |
5.4 |
GO:0060666 |
dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.7 |
2.2 |
GO:0007521 |
muscle cell fate determination(GO:0007521) cellular response to parathyroid hormone stimulus(GO:0071374) positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.7 |
2.2 |
GO:0043379 |
memory T cell differentiation(GO:0043379) |
| 0.7 |
2.1 |
GO:0090258 |
negative regulation of mitochondrial fission(GO:0090258) |
| 0.7 |
4.2 |
GO:0035989 |
tendon development(GO:0035989) |
| 0.7 |
8.0 |
GO:0060501 |
positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.5 |
3.0 |
GO:0015015 |
heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.5 |
1.4 |
GO:0007621 |
negative regulation of female receptivity(GO:0007621) |
| 0.4 |
3.1 |
GO:0051461 |
protein import into peroxisome matrix, docking(GO:0016560) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.4 |
0.9 |
GO:0072197 |
ureter morphogenesis(GO:0072197) |
| 0.4 |
1.3 |
GO:0007341 |
penetration of zona pellucida(GO:0007341) |
| 0.4 |
1.3 |
GO:0051933 |
amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.4 |
2.5 |
GO:0003406 |
retinal pigment epithelium development(GO:0003406) |
| 0.4 |
6.9 |
GO:0032060 |
bleb assembly(GO:0032060) |
| 0.4 |
1.1 |
GO:0032918 |
polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.4 |
1.1 |
GO:0033364 |
mast cell secretory granule organization(GO:0033364) |
| 0.4 |
4.3 |
GO:0061299 |
retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.3 |
1.0 |
GO:0003278 |
apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.3 |
1.6 |
GO:0070295 |
renal water absorption(GO:0070295) |
| 0.3 |
1.9 |
GO:0007320 |
insemination(GO:0007320) |
| 0.3 |
1.8 |
GO:0060024 |
rhythmic synaptic transmission(GO:0060024) |
| 0.3 |
1.1 |
GO:0072488 |
ammonium transmembrane transport(GO:0072488) |
| 0.3 |
2.2 |
GO:0032808 |
lacrimal gland development(GO:0032808) |
| 0.3 |
2.7 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 0.2 |
1.2 |
GO:0015788 |
UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.2 |
4.6 |
GO:0031115 |
negative regulation of microtubule polymerization(GO:0031115) |
| 0.2 |
0.6 |
GO:0045409 |
negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.2 |
1.4 |
GO:2000807 |
regulation of synaptic vesicle clustering(GO:2000807) |
| 0.2 |
1.0 |
GO:1903361 |
protein localization to basolateral plasma membrane(GO:1903361) |
| 0.2 |
1.5 |
GO:0048702 |
embryonic neurocranium morphogenesis(GO:0048702) |
| 0.2 |
0.7 |
GO:0035521 |
monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 |
1.4 |
GO:1902969 |
mitotic DNA replication(GO:1902969) |
| 0.1 |
1.0 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) |
| 0.1 |
1.2 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 |
2.6 |
GO:0002495 |
antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.1 |
0.4 |
GO:0002184 |
cytoplasmic translational termination(GO:0002184) |
| 0.1 |
4.3 |
GO:0030947 |
regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030947) |
| 0.1 |
0.6 |
GO:0071894 |
histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.1 |
0.5 |
GO:0048739 |
cardiac muscle fiber development(GO:0048739) |
| 0.1 |
1.9 |
GO:0072520 |
seminiferous tubule development(GO:0072520) |
| 0.1 |
0.6 |
GO:0046598 |
positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 |
0.7 |
GO:0048934 |
peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 |
1.2 |
GO:0007190 |
activation of adenylate cyclase activity(GO:0007190) |
| 0.1 |
0.9 |
GO:0031339 |
negative regulation of vesicle fusion(GO:0031339) |
| 0.1 |
0.7 |
GO:0071361 |
cellular response to ethanol(GO:0071361) |
| 0.1 |
0.8 |
GO:1904707 |
cellular response to reactive nitrogen species(GO:1902170) positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.1 |
1.2 |
GO:0045954 |
positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.1 |
0.4 |
GO:0006046 |
N-acetylglucosamine catabolic process(GO:0006046) |
| 0.1 |
0.8 |
GO:0060363 |
cranial suture morphogenesis(GO:0060363) |
| 0.1 |
0.7 |
GO:0021521 |
ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 |
0.9 |
GO:0044243 |
collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.1 |
2.1 |
GO:0030199 |
collagen fibril organization(GO:0030199) |
| 0.1 |
0.8 |
GO:0050965 |
detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.1 |
1.5 |
GO:0006491 |
N-glycan processing(GO:0006491) |
| 0.1 |
0.3 |
GO:0051694 |
pointed-end actin filament capping(GO:0051694) |
| 0.0 |
0.2 |
GO:0070427 |
nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 |
0.8 |
GO:0030575 |
nuclear body organization(GO:0030575) |
| 0.0 |
0.9 |
GO:0051894 |
positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 |
0.6 |
GO:0043586 |
tongue development(GO:0043586) |
| 0.0 |
0.1 |
GO:0001787 |
natural killer cell proliferation(GO:0001787) |
| 0.0 |
762.8 |
GO:0008150 |
biological_process(GO:0008150) |