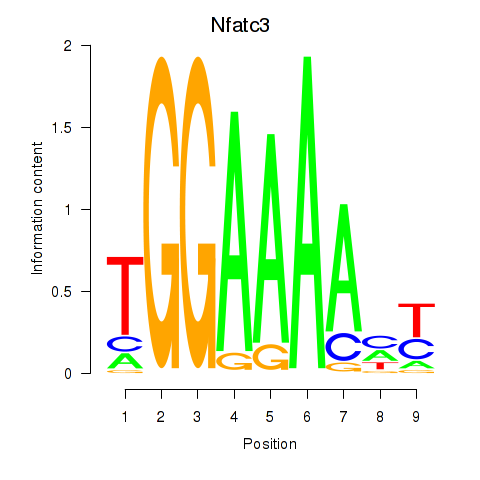
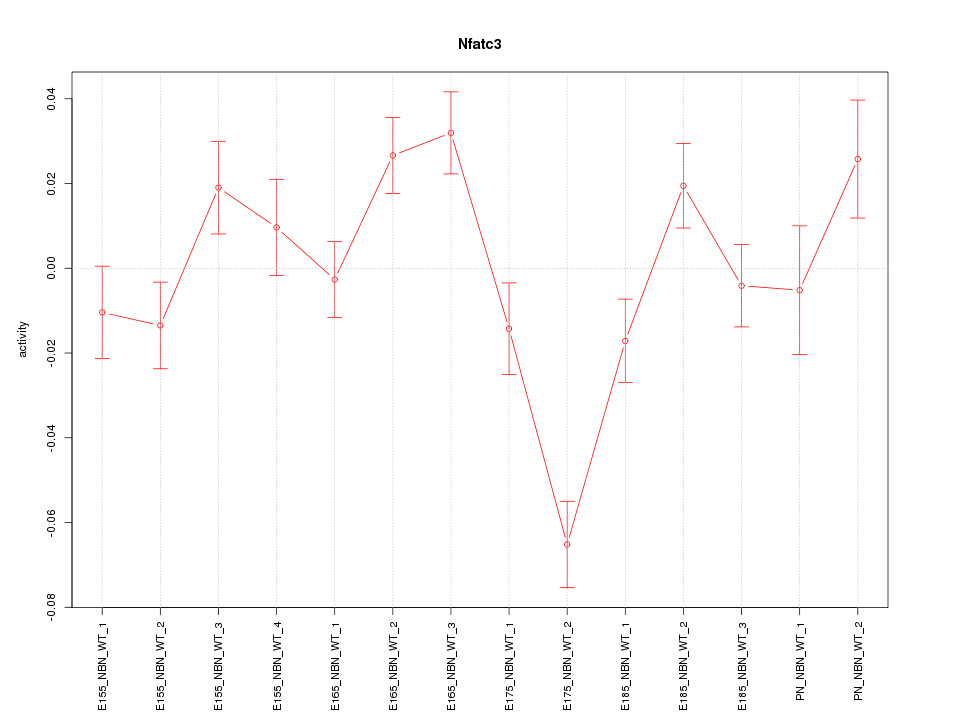
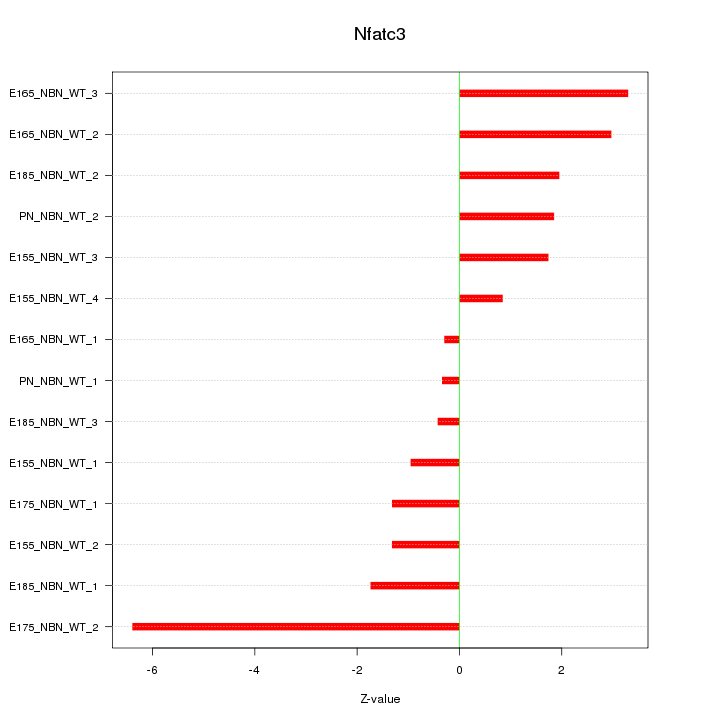
| 2.7 |
8.0 |
GO:0002302 |
CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 1.0 |
7.0 |
GO:2001199 |
negative regulation of dendritic cell differentiation(GO:2001199) |
| 1.0 |
2.9 |
GO:0060854 |
patterning of lymph vessels(GO:0060854) |
| 0.9 |
2.8 |
GO:0003278 |
apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.7 |
3.0 |
GO:0014846 |
esophagus smooth muscle contraction(GO:0014846) |
| 0.7 |
2.8 |
GO:0046013 |
regulation of T cell homeostatic proliferation(GO:0046013) positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.7 |
2.6 |
GO:0060745 |
mammary gland branching involved in pregnancy(GO:0060745) |
| 0.6 |
6.9 |
GO:0061179 |
negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.5 |
3.0 |
GO:0021797 |
forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.5 |
1.4 |
GO:0032918 |
polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.5 |
1.4 |
GO:1903903 |
striated muscle atrophy(GO:0014891) positive regulation of keratinocyte apoptotic process(GO:1902174) regulation of establishment of T cell polarity(GO:1903903) |
| 0.4 |
2.2 |
GO:0003406 |
retinal pigment epithelium development(GO:0003406) |
| 0.4 |
5.5 |
GO:0060159 |
regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.4 |
0.4 |
GO:1905005 |
regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905005) |
| 0.4 |
1.2 |
GO:2000297 |
negative regulation of synapse maturation(GO:2000297) |
| 0.4 |
2.7 |
GO:0072539 |
T-helper 17 cell differentiation(GO:0072539) |
| 0.4 |
2.1 |
GO:0021730 |
trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) |
| 0.3 |
1.0 |
GO:0014877 |
response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.3 |
1.7 |
GO:0010991 |
negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.3 |
1.6 |
GO:0071205 |
clustering of voltage-gated potassium channels(GO:0045163) protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.3 |
0.9 |
GO:0043696 |
dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.3 |
0.9 |
GO:0072554 |
arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) endothelial cell fate specification(GO:0060847) Notch signaling pathway involved in arterial endothelial cell fate commitment(GO:0060853) blood vessel lumenization(GO:0072554) blood vessel endothelial cell fate specification(GO:0097101) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of heart induction(GO:1901321) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.3 |
1.5 |
GO:0019659 |
lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.3 |
0.9 |
GO:1901552 |
positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.3 |
1.2 |
GO:0036438 |
maintenance of lens transparency(GO:0036438) |
| 0.3 |
0.8 |
GO:1900158 |
negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.3 |
1.0 |
GO:0048539 |
bone marrow development(GO:0048539) |
| 0.2 |
2.1 |
GO:0042760 |
very long-chain fatty acid catabolic process(GO:0042760) |
| 0.2 |
1.6 |
GO:0090091 |
positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.2 |
0.6 |
GO:0033092 |
positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.2 |
1.5 |
GO:0034393 |
positive regulation of smooth muscle cell apoptotic process(GO:0034393) follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.2 |
1.2 |
GO:0061365 |
positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.2 |
1.0 |
GO:0060838 |
lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.2 |
0.2 |
GO:0042758 |
long-chain fatty acid catabolic process(GO:0042758) |
| 0.2 |
0.6 |
GO:2000977 |
inhibition of neuroepithelial cell differentiation(GO:0002085) oculomotor nerve development(GO:0021557) ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) negative regulation of auditory receptor cell differentiation(GO:0045608) regulation of forebrain neuron differentiation(GO:2000977) |
| 0.2 |
0.6 |
GO:0046671 |
regulation of cellular pH reduction(GO:0032847) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.2 |
0.6 |
GO:0007290 |
spermatid nucleus elongation(GO:0007290) |
| 0.2 |
1.8 |
GO:0051574 |
positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.2 |
1.5 |
GO:0043568 |
positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.2 |
1.0 |
GO:0043137 |
DNA replication, removal of RNA primer(GO:0043137) |
| 0.2 |
1.6 |
GO:0032464 |
positive regulation of protein homooligomerization(GO:0032464) |
| 0.2 |
0.8 |
GO:0089700 |
positive regulation of T cell receptor signaling pathway(GO:0050862) protein kinase D signaling(GO:0089700) |
| 0.1 |
1.2 |
GO:0035881 |
amacrine cell differentiation(GO:0035881) |
| 0.1 |
0.4 |
GO:1902396 |
protein localization to bicellular tight junction(GO:1902396) |
| 0.1 |
1.2 |
GO:0002536 |
respiratory burst involved in inflammatory response(GO:0002536) |
| 0.1 |
0.4 |
GO:0097309 |
cap1 mRNA methylation(GO:0097309) |
| 0.1 |
0.4 |
GO:0001907 |
killing by symbiont of host cells(GO:0001907) induction of programmed cell death(GO:0012502) disruption by symbiont of host cell(GO:0044004) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.1 |
0.8 |
GO:1901837 |
negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.1 |
0.6 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |
| 0.1 |
0.8 |
GO:2001269 |
positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.1 |
0.6 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.1 |
3.5 |
GO:0040015 |
negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 |
0.9 |
GO:0046645 |
positive regulation of gamma-delta T cell differentiation(GO:0045588) positive regulation of gamma-delta T cell activation(GO:0046645) positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.1 |
0.6 |
GO:0032911 |
nerve growth factor production(GO:0032902) negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 |
0.3 |
GO:0035021 |
negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 |
0.7 |
GO:0006363 |
termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 |
1.5 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 |
0.2 |
GO:0003256 |
regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003256) |
| 0.1 |
0.4 |
GO:0035063 |
nuclear speck organization(GO:0035063) |
| 0.1 |
1.7 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 |
1.4 |
GO:0051451 |
myoblast migration(GO:0051451) |
| 0.1 |
0.5 |
GO:0048739 |
cardiac muscle fiber development(GO:0048739) |
| 0.1 |
0.4 |
GO:2001170 |
negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.1 |
1.0 |
GO:0030575 |
nuclear body organization(GO:0030575) |
| 0.1 |
0.2 |
GO:0070543 |
response to linoleic acid(GO:0070543) |
| 0.1 |
0.4 |
GO:0009169 |
purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.1 |
0.3 |
GO:0070425 |
negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 0.1 |
1.4 |
GO:0032981 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 |
0.3 |
GO:0042905 |
9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 |
0.6 |
GO:0035520 |
monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.1 |
0.8 |
GO:1900119 |
positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.1 |
0.9 |
GO:0097688 |
AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.1 |
0.6 |
GO:0071447 |
cellular response to hydroperoxide(GO:0071447) |
| 0.1 |
0.5 |
GO:0001887 |
selenium compound metabolic process(GO:0001887) |
| 0.1 |
0.2 |
GO:0009838 |
abscission(GO:0009838) |
| 0.1 |
1.0 |
GO:0030953 |
astral microtubule organization(GO:0030953) |
| 0.1 |
0.6 |
GO:0046485 |
ether lipid metabolic process(GO:0046485) |
| 0.1 |
0.6 |
GO:0002755 |
MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.1 |
0.4 |
GO:2000491 |
positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 |
0.3 |
GO:0002922 |
positive regulation of humoral immune response(GO:0002922) progesterone receptor signaling pathway(GO:0050847) |
| 0.1 |
0.6 |
GO:0000963 |
mitochondrial RNA processing(GO:0000963) |
| 0.1 |
1.0 |
GO:0006744 |
ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.1 |
0.9 |
GO:0050860 |
negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 |
0.2 |
GO:0006662 |
glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.1 |
2.0 |
GO:0050819 |
negative regulation of coagulation(GO:0050819) |
| 0.1 |
0.3 |
GO:0060689 |
cell differentiation involved in salivary gland development(GO:0060689) |
| 0.1 |
0.9 |
GO:0007530 |
sex determination(GO:0007530) |
| 0.1 |
0.3 |
GO:1904263 |
positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 |
1.1 |
GO:0010667 |
negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.1 |
0.5 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 |
0.6 |
GO:0010447 |
response to acidic pH(GO:0010447) |
| 0.0 |
0.8 |
GO:0032060 |
bleb assembly(GO:0032060) |
| 0.0 |
0.3 |
GO:0006287 |
base-excision repair, gap-filling(GO:0006287) |
| 0.0 |
0.4 |
GO:0031339 |
negative regulation of vesicle fusion(GO:0031339) |
| 0.0 |
0.4 |
GO:0071225 |
cellular response to muramyl dipeptide(GO:0071225) |
| 0.0 |
0.2 |
GO:1904749 |
regulation of protein localization to nucleolus(GO:1904749) positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.0 |
0.2 |
GO:0045578 |
negative regulation of B cell differentiation(GO:0045578) |
| 0.0 |
0.6 |
GO:0044406 |
adhesion of symbiont to host(GO:0044406) |
| 0.0 |
0.5 |
GO:0050892 |
intestinal absorption(GO:0050892) |
| 0.0 |
0.2 |
GO:0032808 |
lacrimal gland development(GO:0032808) |
| 0.0 |
0.5 |
GO:1901898 |
negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 |
0.4 |
GO:0006415 |
translational termination(GO:0006415) |
| 0.0 |
1.7 |
GO:0045600 |
positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 |
0.3 |
GO:0021869 |
forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 |
0.2 |
GO:1903352 |
ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.0 |
0.2 |
GO:0061087 |
positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 |
0.3 |
GO:0035970 |
peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 |
0.4 |
GO:0043586 |
tongue development(GO:0043586) |
| 0.0 |
0.6 |
GO:0009950 |
dorsal/ventral axis specification(GO:0009950) |
| 0.0 |
1.3 |
GO:0060325 |
face morphogenesis(GO:0060325) |
| 0.0 |
0.3 |
GO:0060394 |
negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 |
0.2 |
GO:0030579 |
ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 |
1.1 |
GO:0021517 |
ventral spinal cord development(GO:0021517) |
| 0.0 |
1.7 |
GO:0051489 |
regulation of filopodium assembly(GO:0051489) |
| 0.0 |
1.7 |
GO:0032024 |
positive regulation of insulin secretion(GO:0032024) |
| 0.0 |
0.1 |
GO:1901301 |
regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 |
3.1 |
GO:0051028 |
mRNA transport(GO:0051028) |
| 0.0 |
0.2 |
GO:0021814 |
cell motility involved in cerebral cortex radial glia guided migration(GO:0021814) |
| 0.0 |
0.2 |
GO:0016446 |
somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 |
0.1 |
GO:0035356 |
cellular triglyceride homeostasis(GO:0035356) adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) |
| 0.0 |
0.2 |
GO:0097119 |
postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 |
0.3 |
GO:0010758 |
regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 |
0.1 |
GO:0015014 |
heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) ureteric bud formation(GO:0060676) |
| 0.0 |
0.1 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.0 |
0.3 |
GO:0036010 |
protein localization to endosome(GO:0036010) |
| 0.0 |
1.3 |
GO:0001578 |
microtubule bundle formation(GO:0001578) |
| 0.0 |
0.2 |
GO:2000353 |
positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 |
0.4 |
GO:0021904 |
dorsal/ventral neural tube patterning(GO:0021904) |
| 0.0 |
0.6 |
GO:0015804 |
neutral amino acid transport(GO:0015804) |
| 0.0 |
0.3 |
GO:0016556 |
mRNA modification(GO:0016556) |
| 0.0 |
2.0 |
GO:0035023 |
regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 |
0.1 |
GO:0000160 |
phosphorelay signal transduction system(GO:0000160) |
| 0.0 |
0.1 |
GO:0097011 |
cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.0 |
0.2 |
GO:0051894 |
positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 |
0.3 |
GO:0061178 |
regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 |
0.1 |
GO:0097298 |
regulation of nucleus size(GO:0097298) |
| 0.0 |
0.9 |
GO:0032526 |
response to retinoic acid(GO:0032526) |
| 0.0 |
0.1 |
GO:0045065 |
cytotoxic T cell differentiation(GO:0045065) |
| 0.0 |
0.2 |
GO:0060009 |
Sertoli cell development(GO:0060009) |
| 0.0 |
0.4 |
GO:0046470 |
phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 |
0.1 |
GO:0021993 |
initiation of neural tube closure(GO:0021993) |
| 0.0 |
0.3 |
GO:0003301 |
physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 |
0.3 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.0 |
0.1 |
GO:0030836 |
positive regulation of actin filament depolymerization(GO:0030836) |
| 0.0 |
0.5 |
GO:0006506 |
GPI anchor biosynthetic process(GO:0006506) |