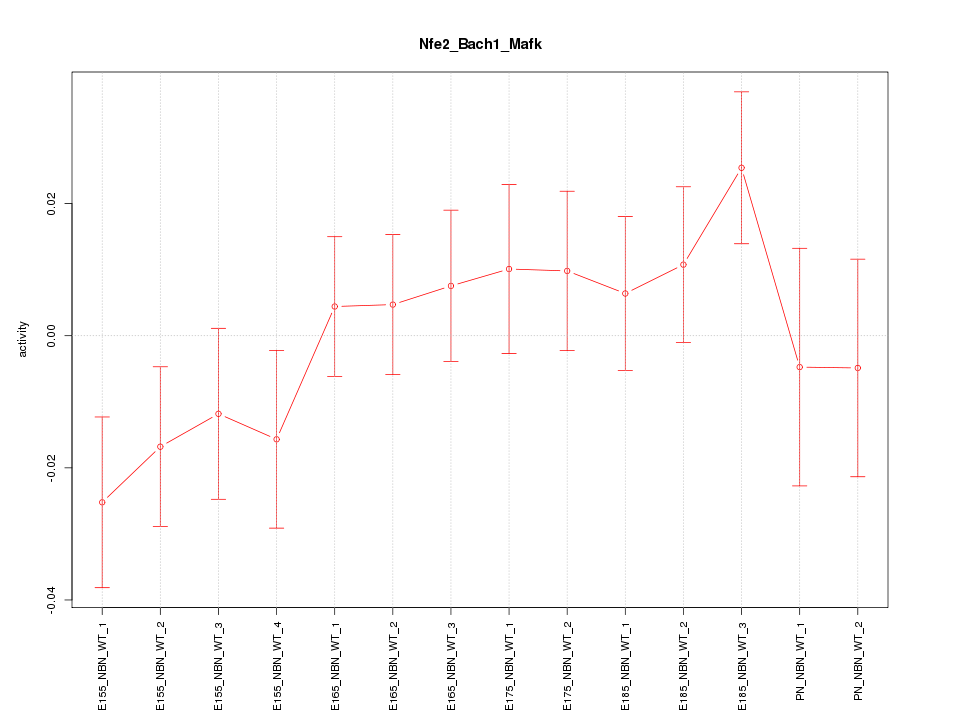
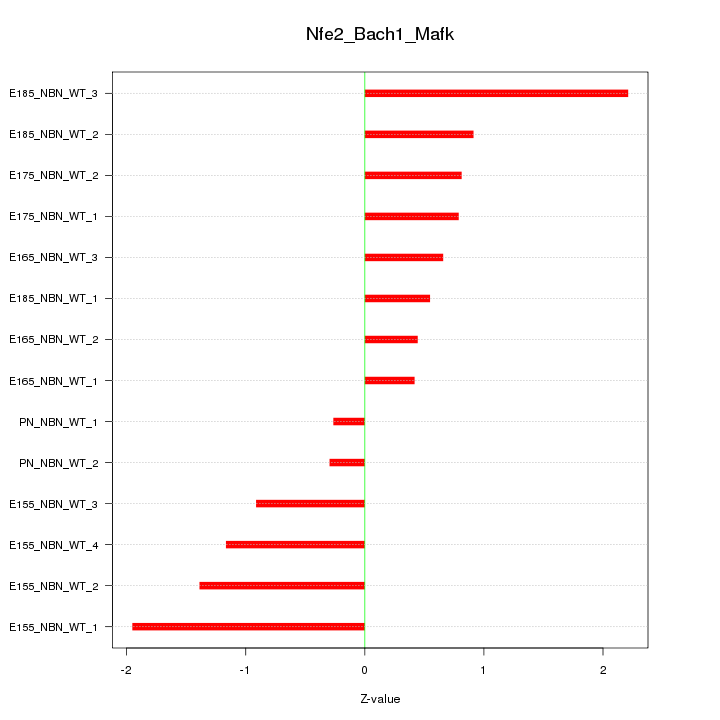
Motif ID: Nfe2_Bach1_Mafk
Z-value: 1.076
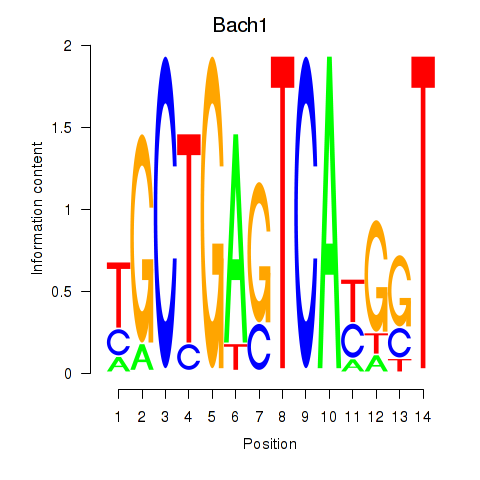
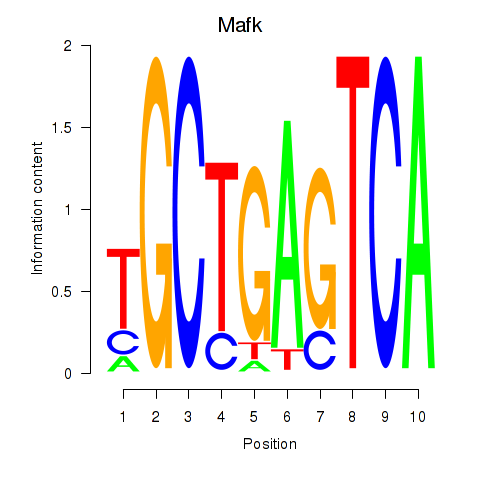
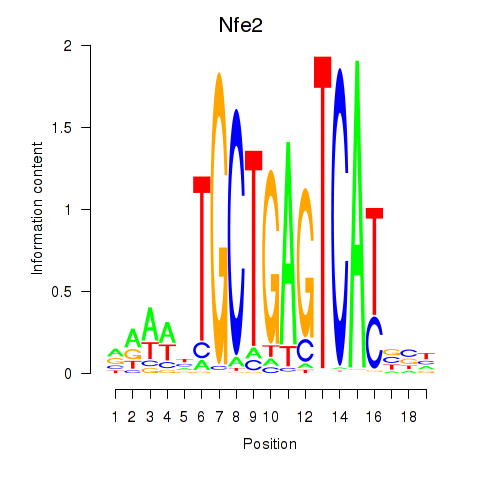
Transcription factors associated with Nfe2_Bach1_Mafk:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Bach1 | ENSMUSG00000025612.5 | Bach1 |
| Mafk | ENSMUSG00000018143.4 | Mafk |
| Nfe2 | ENSMUSG00000058794.6 | Nfe2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Bach1 | mm10_v2_chr16_+_87698904_87698959 | -0.49 | 7.6e-02 | Click! |
| Nfe2 | mm10_v2_chr15_-_103251465_103251705 | -0.26 | 3.7e-01 | Click! |
| Mafk | mm10_v2_chr5_+_139791513_139791539 | -0.11 | 7.1e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 5.3 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.4 | 1.8 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.3 | 1.0 | GO:0090272 | negative regulation of fibroblast growth factor production(GO:0090272) |
| 0.3 | 0.5 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.2 | 1.9 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.2 | 1.2 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.2 | 1.6 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.2 | 0.6 | GO:2000501 | natural killer cell chemotaxis(GO:0035747) negative regulation of lymphocyte migration(GO:2000402) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.2 | 0.4 | GO:0043173 | nucleotide salvage(GO:0043173) |
| 0.2 | 0.5 | GO:0070488 | neutrophil aggregation(GO:0070488) |
| 0.2 | 0.5 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.2 | 1.3 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.2 | 4.2 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.2 | 0.2 | GO:0031118 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) rRNA pseudouridine synthesis(GO:0031118) |
| 0.1 | 0.6 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 0.4 | GO:0002578 | negative regulation of antigen processing and presentation(GO:0002578) |
| 0.1 | 1.3 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.1 | 1.0 | GO:0009181 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.1 | 0.6 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.1 | 0.3 | GO:0072204 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.1 | 2.1 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 0.6 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.1 | 0.5 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.1 | 0.7 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 1.2 | GO:0010819 | regulation of T cell chemotaxis(GO:0010819) |
| 0.1 | 0.3 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.1 | 0.9 | GO:0044331 | cell-cell adhesion mediated by cadherin(GO:0044331) |
| 0.1 | 0.4 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.1 | 0.3 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) metanephric glomerular mesangium development(GO:0072223) metanephric glomerular mesangial cell proliferation involved in metanephros development(GO:0072262) |
| 0.1 | 0.3 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.1 | 0.2 | GO:0043465 | regulation of fermentation(GO:0043465) negative regulation of fermentation(GO:1901003) |
| 0.1 | 0.8 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 0.6 | GO:2000809 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.1 | 0.3 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 0.4 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.1 | 0.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.5 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 0.1 | GO:0002295 | T-helper cell lineage commitment(GO:0002295) |
| 0.1 | 0.2 | GO:0043323 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.1 | 0.3 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.1 | 0.2 | GO:0001978 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) |
| 0.1 | 0.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 0.5 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.4 | GO:0061620 | glycolytic process through glucose-6-phosphate(GO:0061620) |
| 0.0 | 0.9 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.2 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.0 | 0.6 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.3 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.4 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0071451 | cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) |
| 0.0 | 0.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.2 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.2 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.0 | 0.1 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.3 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.4 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.1 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.6 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.0 | 1.0 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 0.1 | GO:2000195 | negative regulation of female gonad development(GO:2000195) |
| 0.0 | 0.1 | GO:0060745 | mammary gland branching involved in pregnancy(GO:0060745) |
| 0.0 | 0.2 | GO:1903847 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:0070305 | response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) |
| 0.0 | 0.1 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.0 | GO:2001185 | regulation of CD8-positive, alpha-beta T cell activation(GO:2001185) |
| 0.0 | 0.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.3 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.1 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 0.1 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.0 | 0.1 | GO:1903423 | positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 1.1 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.1 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.0 | 0.1 | GO:0090274 | positive regulation of somatostatin secretion(GO:0090274) |
| 0.0 | 0.1 | GO:0007603 | phototransduction, visible light(GO:0007603) |
| 0.0 | 0.1 | GO:0046103 | ADP biosynthetic process(GO:0006172) inosine biosynthetic process(GO:0046103) |
| 0.0 | 0.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.6 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.3 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.2 | GO:2000780 | negative regulation of double-strand break repair(GO:2000780) |
| 0.0 | 0.9 | GO:0030641 | regulation of cellular pH(GO:0030641) |
| 0.0 | 0.1 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.0 | 0.2 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.0 | GO:0070589 | cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) cell wall biogenesis(GO:0042546) cell wall macromolecule metabolic process(GO:0044036) cell wall macromolecule biosynthetic process(GO:0044038) chain elongation of O-linked mannose residue(GO:0044845) cellular component macromolecule biosynthetic process(GO:0070589) cell wall organization or biogenesis(GO:0071554) |
| 0.0 | 0.5 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.1 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.0 | GO:0021506 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.0 | 0.5 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.3 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.2 | 0.6 | GO:0036501 | UFD1-NPL4 complex(GO:0036501) |
| 0.2 | 0.5 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.2 | 0.5 | GO:0044754 | amphisome(GO:0044753) autolysosome(GO:0044754) |
| 0.1 | 3.3 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.6 | GO:1990462 | omegasome(GO:1990462) |
| 0.1 | 0.6 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 0.4 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 0.5 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 0.7 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.1 | 4.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.2 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.1 | 0.8 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 0.3 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 1.0 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 0.5 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.6 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.7 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.6 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.2 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.2 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 2.0 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.5 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.4 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 1.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.2 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.8 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.1 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.5 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.9 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.2 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.1 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.6 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 1.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.4 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.3 | 1.9 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.3 | 3.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.2 | 0.9 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.6 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 0.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 2.2 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 1.0 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.1 | 1.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.3 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.6 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.1 | 0.9 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.2 | GO:0004935 | adrenergic receptor activity(GO:0004935) alpha-adrenergic receptor activity(GO:0004936) |
| 0.1 | 0.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.6 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 1.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.7 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.3 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.1 | 0.3 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 0.5 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.4 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 0.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.6 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.1 | 0.2 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 0.6 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 0.3 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.1 | 0.4 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.4 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.3 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.0 | 1.1 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.9 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.7 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.1 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.2 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 1.0 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 1.5 | GO:0016860 | intramolecular oxidoreductase activity(GO:0016860) |
| 0.0 | 0.1 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.2 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.3 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.9 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.1 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.0 | 0.4 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.3 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.7 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.9 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.4 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.5 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.1 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.3 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 1.0 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.0 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |