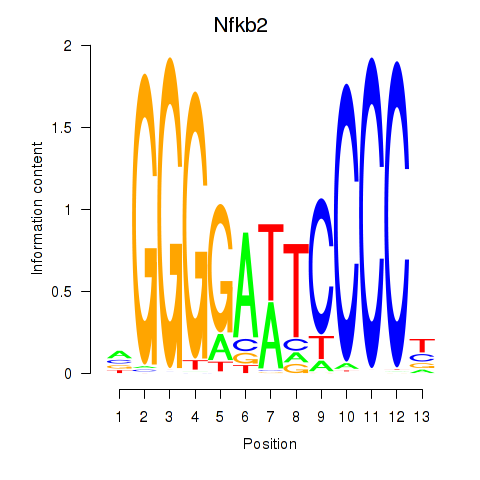
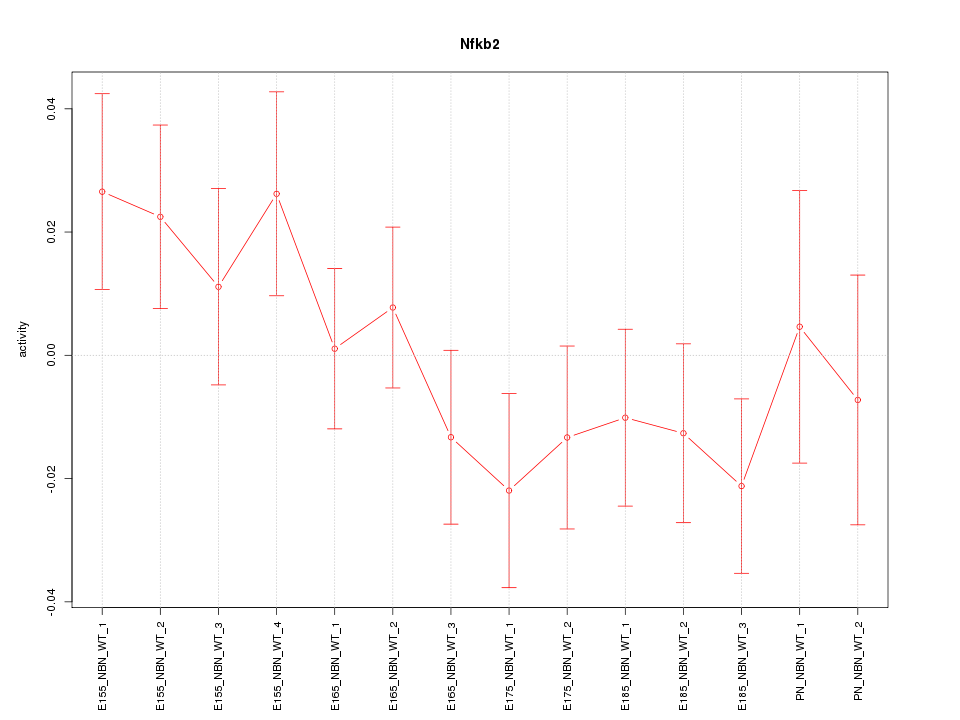
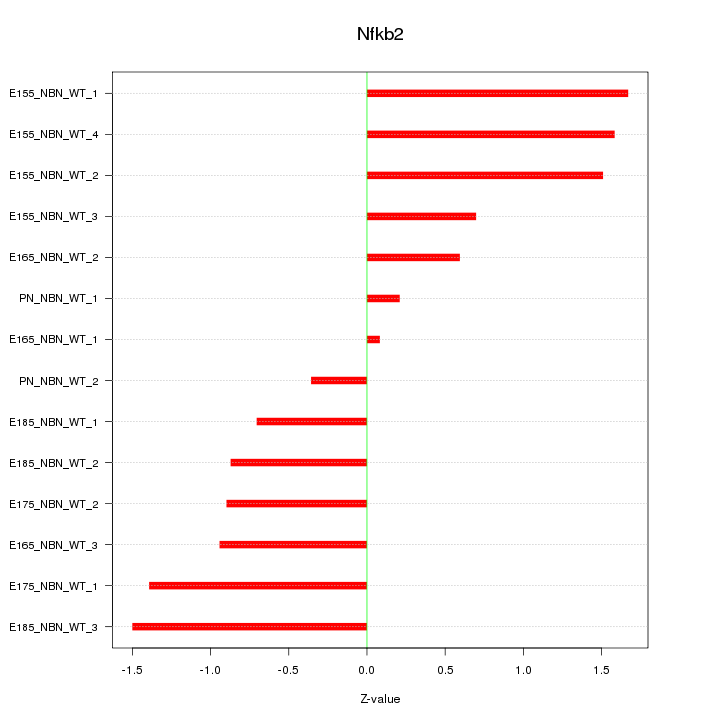
| 1.7 |
6.7 |
GO:0070976 |
calcium-independent protein kinase C activity(GO:0004699) TIR domain binding(GO:0070976) |
| 1.5 |
4.6 |
GO:0005105 |
type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.2 |
1.9 |
GO:0004865 |
protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 |
0.2 |
GO:0017089 |
glycolipid transporter activity(GO:0017089) |
| 0.1 |
0.3 |
GO:0008349 |
MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 |
4.1 |
GO:0000980 |
RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 |
0.7 |
GO:0003708 |
retinoic acid receptor activity(GO:0003708) |
| 0.0 |
1.6 |
GO:0005109 |
frizzled binding(GO:0005109) |
| 0.0 |
0.4 |
GO:0035925 |
mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 |
0.3 |
GO:0004634 |
phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 |
3.0 |
GO:0008565 |
protein transporter activity(GO:0008565) |
| 0.0 |
0.5 |
GO:0004679 |
AMP-activated protein kinase activity(GO:0004679) |
| 0.0 |
1.1 |
GO:0051019 |
mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 |
1.4 |
GO:0004722 |
protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 |
5.5 |
GO:0005096 |
GTPase activator activity(GO:0005096) |
| 0.0 |
0.2 |
GO:0030881 |
beta-2-microglobulin binding(GO:0030881) |
| 0.0 |
0.9 |
GO:0035496 |
UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) O antigen polymerase activity(GO:0008755) lipopolysaccharide-1,6-galactosyltransferase activity(GO:0008921) cellulose synthase activity(GO:0016759) 9-phenanthrol UDP-glucuronosyltransferase activity(GO:0018715) 1-phenanthrol glycosyltransferase activity(GO:0018716) 9-phenanthrol glycosyltransferase activity(GO:0018717) 1,2-dihydroxy-phenanthrene glycosyltransferase activity(GO:0018718) phenanthrol glycosyltransferase activity(GO:0019112) alpha-1,2-galactosyltransferase activity(GO:0031278) dolichyl pyrophosphate Man7GlcNAc2 alpha-1,3-glucosyltransferase activity(GO:0033556) endogalactosaminidase activity(GO:0033931) lipopolysaccharide-1,5-galactosyltransferase activity(GO:0035496) dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase activity(GO:0042283) inositol phosphoceramide synthase activity(GO:0045140) alpha-(1->3)-fucosyltransferase activity(GO:0046920) indole-3-butyrate beta-glucosyltransferase activity(GO:0052638) salicylic acid glucosyltransferase (ester-forming) activity(GO:0052639) salicylic acid glucosyltransferase (glucoside-forming) activity(GO:0052640) benzoic acid glucosyltransferase activity(GO:0052641) chondroitin hydrolase activity(GO:0052757) dolichyl-pyrophosphate Man7GlcNAc2 alpha-1,6-mannosyltransferase activity(GO:0052824) cytokinin 9-beta-glucosyltransferase activity(GO:0080062) |
| 0.0 |
0.2 |
GO:0042556 |
cobinamide kinase activity(GO:0008819) phytol kinase activity(GO:0010276) phenol kinase activity(GO:0018720) cyclin-dependent protein kinase activating kinase regulator activity(GO:0019914) inositol tetrakisphosphate 2-kinase activity(GO:0032942) heptose 7-phosphate kinase activity(GO:0033785) aminoglycoside phosphotransferase activity(GO:0034071) eukaryotic elongation factor-2 kinase regulator activity(GO:0042556) eukaryotic elongation factor-2 kinase activator activity(GO:0042557) LPPG:FO 2-phospho-L-lactate transferase activity(GO:0043743) cytidine kinase activity(GO:0043771) glycerate 2-kinase activity(GO:0043798) (S)-lactate 2-kinase activity(GO:0043841) phosphoserine:homoserine phosphotransferase activity(GO:0043899) L-seryl-tRNA(Sec) kinase activity(GO:0043915) phosphocholine transferase activity(GO:0044605) GTP-dependent polynucleotide kinase activity(GO:0051735) farnesol kinase activity(GO:0052668) CTP:2-trans,-6-trans-farnesol kinase activity(GO:0052669) geraniol kinase activity(GO:0052670) geranylgeraniol kinase activity(GO:0052671) CTP:geranylgeraniol kinase activity(GO:0052672) prenol kinase activity(GO:0052673) 1-phosphatidylinositol-5-kinase activity(GO:0052810) 1-phosphatidylinositol-3-phosphate 4-kinase activity(GO:0052811) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 |
0.5 |
GO:0030291 |
protein serine/threonine kinase inhibitor activity(GO:0030291) |
| 0.0 |
0.1 |
GO:0031749 |
D2 dopamine receptor binding(GO:0031749) |
| 0.0 |
0.3 |
GO:0030676 |
Rac guanyl-nucleotide exchange factor activity(GO:0030676) |