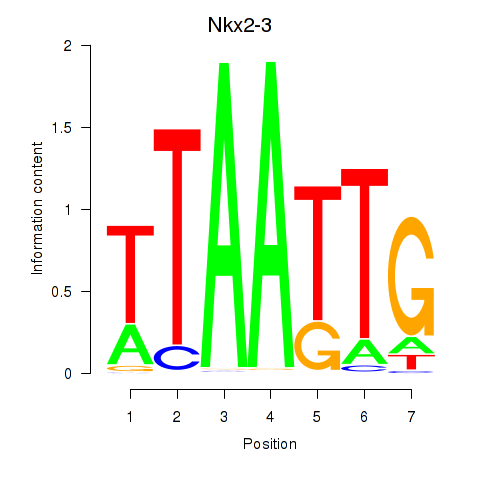
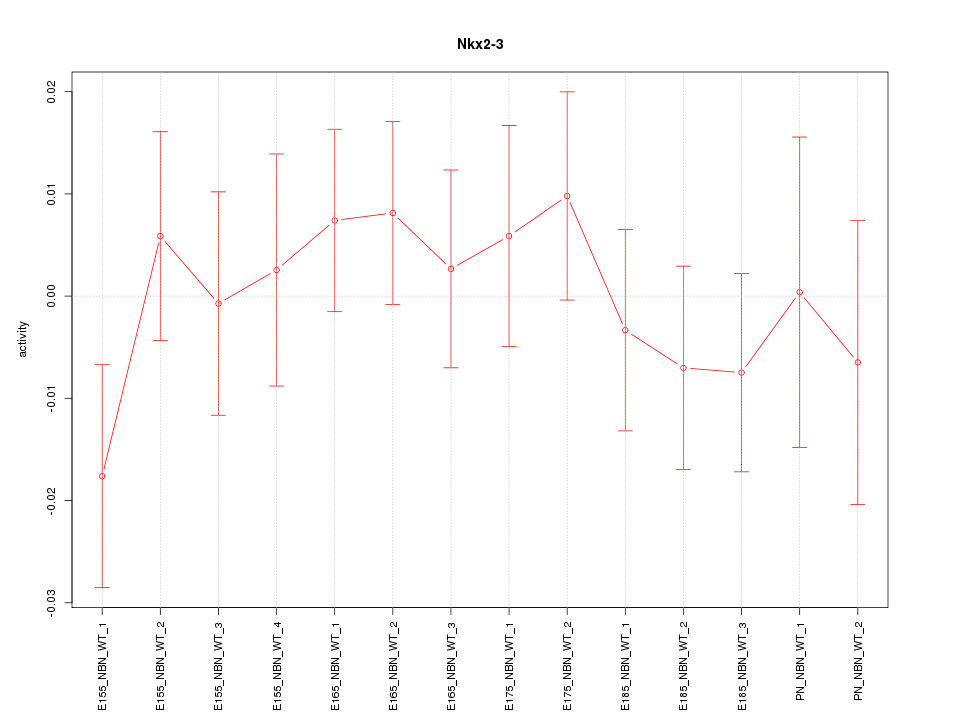
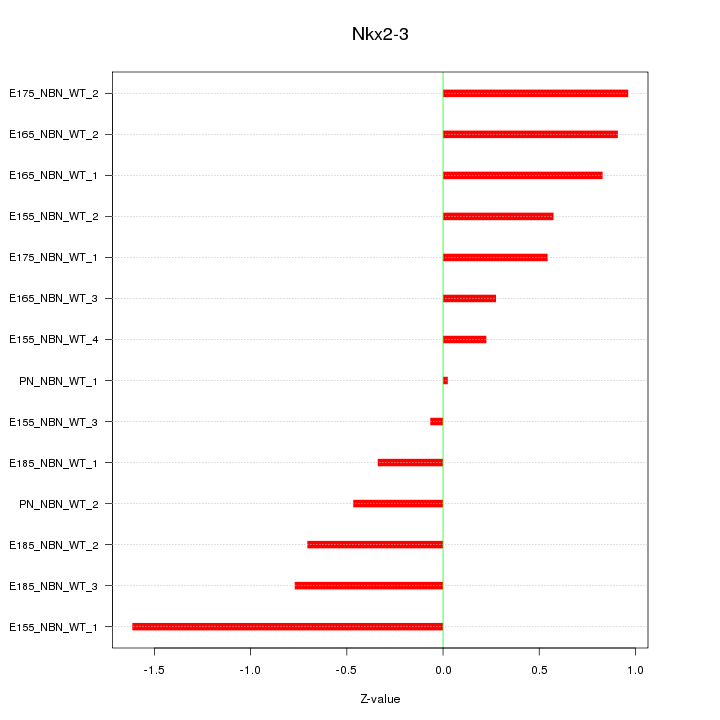
| 0.2 |
0.6 |
GO:0007521 |
muscle cell fate determination(GO:0007521) cellular response to parathyroid hormone stimulus(GO:0071374) positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.2 |
1.2 |
GO:0032625 |
interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.2 |
0.5 |
GO:0050925 |
negative regulation of negative chemotaxis(GO:0050925) |
| 0.1 |
1.1 |
GO:0097264 |
self proteolysis(GO:0097264) |
| 0.1 |
0.4 |
GO:0060489 |
establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.1 |
0.6 |
GO:1903215 |
negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.1 |
0.2 |
GO:2000481 |
positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 0.1 |
0.7 |
GO:0006570 |
tyrosine metabolic process(GO:0006570) |
| 0.1 |
0.3 |
GO:0097155 |
fasciculation of sensory neuron axon(GO:0097155) |
| 0.1 |
0.3 |
GO:1901204 |
positive regulation of guanylate cyclase activity(GO:0031284) regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 0.1 |
0.4 |
GO:0002121 |
inter-male aggressive behavior(GO:0002121) |
| 0.1 |
0.2 |
GO:0016095 |
polyprenol catabolic process(GO:0016095) terpenoid catabolic process(GO:0016115) primary alcohol catabolic process(GO:0034310) |
| 0.1 |
0.4 |
GO:0046684 |
response to pyrethroid(GO:0046684) |
| 0.1 |
0.2 |
GO:0021759 |
globus pallidus development(GO:0021759) |
| 0.1 |
0.6 |
GO:0033353 |
S-adenosylmethionine cycle(GO:0033353) |
| 0.1 |
0.8 |
GO:0032926 |
negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 |
0.5 |
GO:0090074 |
negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 |
0.3 |
GO:0036438 |
maintenance of lens transparency(GO:0036438) |
| 0.1 |
0.5 |
GO:0021830 |
interneuron migration from the subpallium to the cortex(GO:0021830) |
| 0.1 |
0.4 |
GO:1902474 |
positive regulation of protein localization to synapse(GO:1902474) |
| 0.1 |
0.2 |
GO:0035864 |
response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.1 |
0.5 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.0 |
0.8 |
GO:0050966 |
detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.0 |
0.4 |
GO:0036371 |
protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 |
0.2 |
GO:0035063 |
nuclear speck organization(GO:0035063) |
| 0.0 |
0.2 |
GO:1903347 |
negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 |
0.3 |
GO:2000301 |
negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 |
0.6 |
GO:0021860 |
pyramidal neuron development(GO:0021860) |
| 0.0 |
0.2 |
GO:0002317 |
plasma cell differentiation(GO:0002317) |
| 0.0 |
0.3 |
GO:0018230 |
peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 |
0.3 |
GO:1903847 |
regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.0 |
0.1 |
GO:0038095 |
positive regulation of mast cell cytokine production(GO:0032765) Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.0 |
0.3 |
GO:0035095 |
behavioral response to nicotine(GO:0035095) |
| 0.0 |
0.1 |
GO:0060690 |
epithelial cell differentiation involved in salivary gland development(GO:0060690) |
| 0.0 |
0.3 |
GO:0035970 |
peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 |
0.3 |
GO:1901898 |
negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 |
0.2 |
GO:0060018 |
astrocyte fate commitment(GO:0060018) |
| 0.0 |
0.1 |
GO:1902915 |
negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 |
0.2 |
GO:0009404 |
toxin metabolic process(GO:0009404) |
| 0.0 |
0.3 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 |
0.2 |
GO:0035887 |
aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 |
0.1 |
GO:0038032 |
termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.0 |
0.3 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 |
0.2 |
GO:1902965 |
regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.0 |
0.1 |
GO:1903690 |
negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 |
0.1 |
GO:1903056 |
regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.0 |
0.2 |
GO:2001256 |
regulation of store-operated calcium entry(GO:2001256) |
| 0.0 |
0.1 |
GO:0046477 |
glycosylceramide catabolic process(GO:0046477) |
| 0.0 |
0.1 |
GO:0001867 |
complement activation, lectin pathway(GO:0001867) |
| 0.0 |
0.1 |
GO:0090188 |
negative regulation of pancreatic juice secretion(GO:0090188) |
| 0.0 |
0.1 |
GO:0060591 |
chondroblast differentiation(GO:0060591) |
| 0.0 |
0.2 |
GO:0034308 |
primary alcohol metabolic process(GO:0034308) |
| 0.0 |
0.2 |
GO:2000270 |
negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 |
0.1 |
GO:0033540 |
fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 |
0.1 |
GO:1903377 |
negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 |
0.5 |
GO:0021952 |
central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 |
0.3 |
GO:0015701 |
bicarbonate transport(GO:0015701) |
| 0.0 |
0.4 |
GO:0006491 |
N-glycan processing(GO:0006491) |
| 0.0 |
0.1 |
GO:0051964 |
negative regulation of synapse assembly(GO:0051964) |
| 0.0 |
1.0 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 |
0.1 |
GO:0070562 |
regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 |
0.2 |
GO:0080111 |
DNA demethylation(GO:0080111) |
| 0.0 |
0.3 |
GO:0071624 |
positive regulation of granulocyte chemotaxis(GO:0071624) positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 |
0.0 |
GO:0071476 |
hypotonic response(GO:0006971) cellular hypotonic response(GO:0071476) |
| 0.0 |
0.1 |
GO:0010650 |
positive regulation of cell communication by electrical coupling(GO:0010650) cellular response to magnesium ion(GO:0071286) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 |
0.1 |
GO:0045218 |
zonula adherens maintenance(GO:0045218) |
| 0.0 |
0.6 |
GO:0021954 |
central nervous system neuron development(GO:0021954) |
| 0.0 |
0.9 |
GO:0051965 |
positive regulation of synapse assembly(GO:0051965) |
| 0.0 |
0.0 |
GO:0009838 |
abscission(GO:0009838) positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 |
0.1 |
GO:0043508 |
negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 |
0.0 |
GO:0090327 |
negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.0 |
0.1 |
GO:0001731 |
formation of translation preinitiation complex(GO:0001731) |