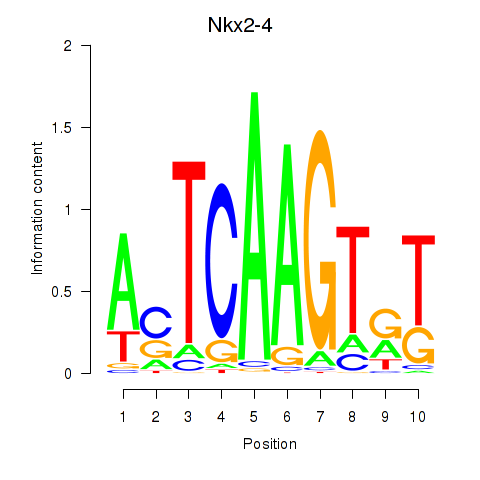
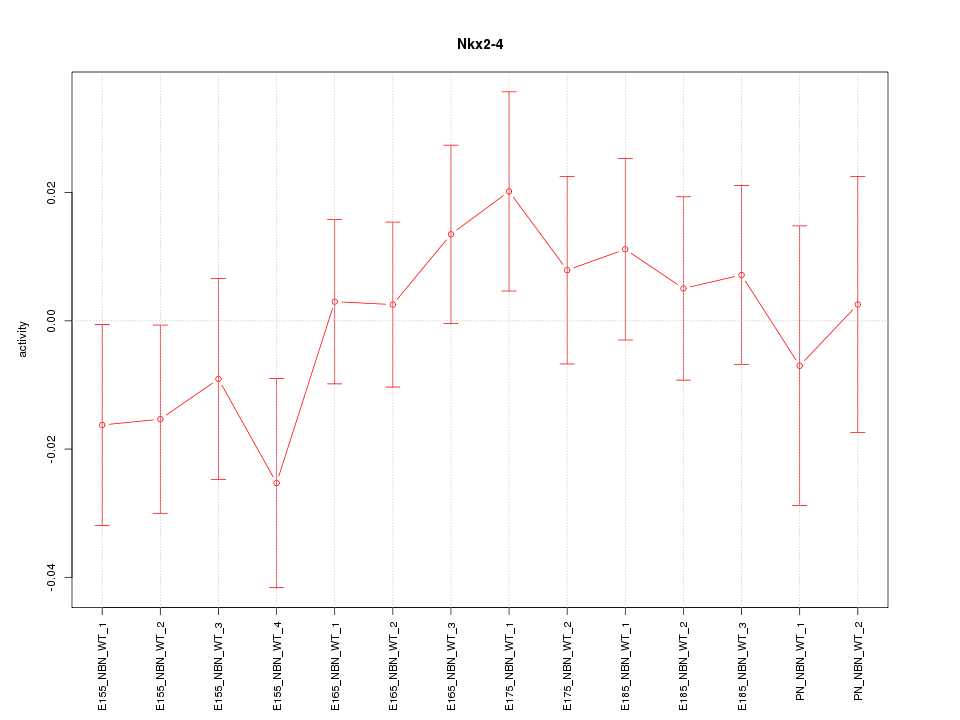
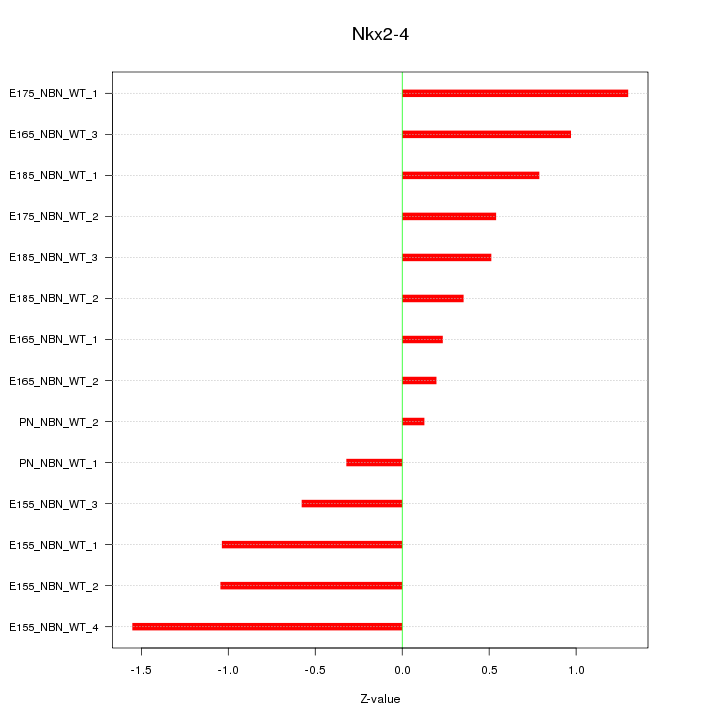
| 1.9 |
5.7 |
GO:0072092 |
ureteric bud invasion(GO:0072092) |
| 0.5 |
3.7 |
GO:0045650 |
negative regulation of macrophage differentiation(GO:0045650) |
| 0.3 |
0.9 |
GO:1904139 |
microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.2 |
1.7 |
GO:0071732 |
cellular response to nitric oxide(GO:0071732) |
| 0.2 |
0.6 |
GO:0021529 |
spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.2 |
0.5 |
GO:1902310 |
positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.1 |
0.3 |
GO:2000642 |
negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.1 |
0.4 |
GO:0006177 |
GMP biosynthetic process(GO:0006177) |
| 0.1 |
0.9 |
GO:0043249 |
erythrocyte maturation(GO:0043249) |
| 0.1 |
1.2 |
GO:0015701 |
bicarbonate transport(GO:0015701) |
| 0.1 |
0.3 |
GO:0051182 |
coenzyme transport(GO:0051182) |
| 0.0 |
1.6 |
GO:0007638 |
mechanosensory behavior(GO:0007638) |
| 0.0 |
1.8 |
GO:0045104 |
intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 |
0.6 |
GO:0042415 |
norepinephrine metabolic process(GO:0042415) |
| 0.0 |
0.8 |
GO:0032981 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 |
0.2 |
GO:0048859 |
formation of anatomical boundary(GO:0048859) |
| 0.0 |
0.1 |
GO:0060745 |
mammary gland branching involved in pregnancy(GO:0060745) |
| 0.0 |
0.1 |
GO:0006049 |
UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 |
1.0 |
GO:0031146 |
SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 |
0.5 |
GO:0033234 |
negative regulation of protein sumoylation(GO:0033234) |
| 0.0 |
0.2 |
GO:0014807 |
regulation of somitogenesis(GO:0014807) |
| 0.0 |
0.4 |
GO:0050832 |
defense response to fungus(GO:0050832) |
| 0.0 |
0.4 |
GO:1904872 |
regulation of telomerase RNA localization to Cajal body(GO:1904872) positive regulation of telomerase RNA localization to Cajal body(GO:1904874) |
| 0.0 |
0.1 |
GO:0010908 |
regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) myoblast fate commitment(GO:0048625) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 |
0.1 |
GO:0070682 |
proteasome regulatory particle assembly(GO:0070682) |
| 0.0 |
0.4 |
GO:0071625 |
vocalization behavior(GO:0071625) |
| 0.0 |
0.1 |
GO:0015868 |
purine ribonucleotide transport(GO:0015868) |
| 0.0 |
0.5 |
GO:0097320 |
membrane tubulation(GO:0097320) |
| 0.0 |
0.3 |
GO:0038092 |
nodal signaling pathway(GO:0038092) |
| 0.0 |
0.3 |
GO:0051127 |
positive regulation of actin nucleation(GO:0051127) |
| 0.0 |
0.2 |
GO:0048280 |
vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 |
0.3 |
GO:0010107 |
potassium ion import(GO:0010107) |