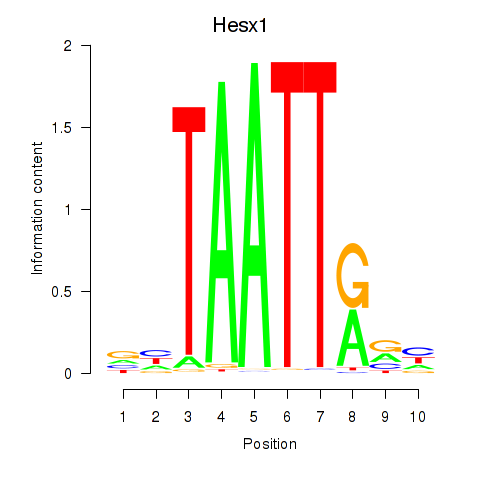
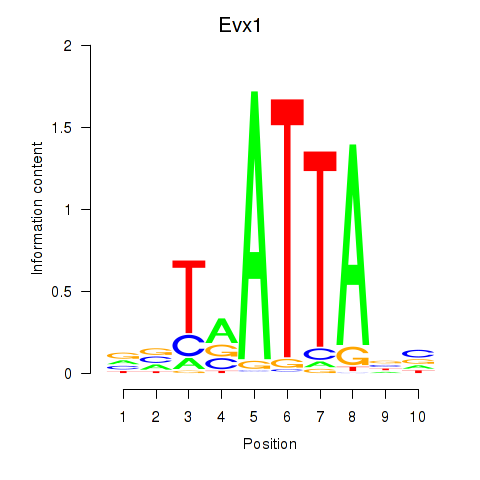
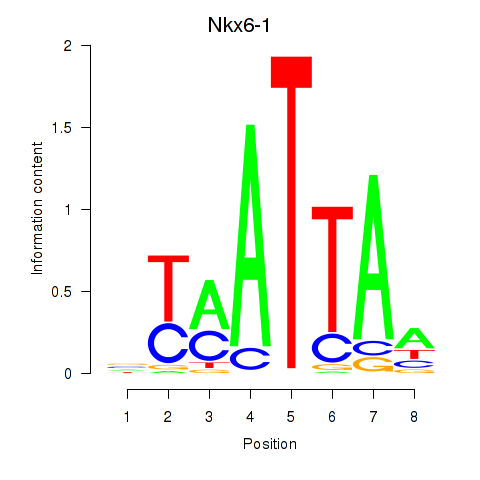
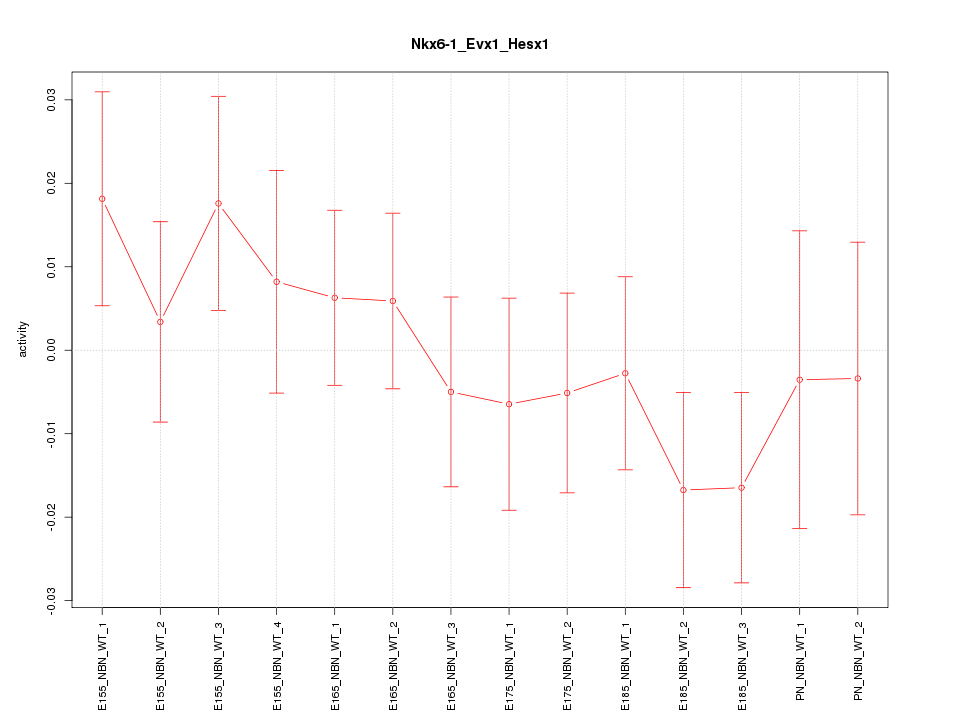
| 1.7 |
5.2 |
GO:0051892 |
negative regulation of cardioblast differentiation(GO:0051892) |
| 1.2 |
3.7 |
GO:0060489 |
establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 1.2 |
3.5 |
GO:0002302 |
CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.8 |
2.4 |
GO:0060478 |
acrosomal vesicle exocytosis(GO:0060478) |
| 0.7 |
4.4 |
GO:0030643 |
cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.4 |
2.6 |
GO:0021797 |
forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.4 |
1.7 |
GO:0071596 |
ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.4 |
1.1 |
GO:0050975 |
sensory perception of touch(GO:0050975) |
| 0.3 |
1.3 |
GO:0014846 |
esophagus smooth muscle contraction(GO:0014846) |
| 0.3 |
1.5 |
GO:0015692 |
vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.3 |
2.9 |
GO:0071372 |
cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.3 |
0.9 |
GO:2000620 |
positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.3 |
1.1 |
GO:0061055 |
myotome development(GO:0061055) |
| 0.3 |
2.3 |
GO:0035726 |
common myeloid progenitor cell proliferation(GO:0035726) |
| 0.3 |
1.4 |
GO:0038032 |
termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.3 |
1.8 |
GO:0031536 |
positive regulation of exit from mitosis(GO:0031536) |
| 0.2 |
0.2 |
GO:0009946 |
proximal/distal axis specification(GO:0009946) |
| 0.2 |
1.4 |
GO:1903056 |
regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.2 |
0.7 |
GO:0042663 |
regulation of endodermal cell fate specification(GO:0042663) |
| 0.2 |
1.0 |
GO:1903690 |
negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 |
3.6 |
GO:0097352 |
autophagosome maturation(GO:0097352) |
| 0.1 |
2.7 |
GO:0021860 |
pyramidal neuron development(GO:0021860) |
| 0.1 |
0.6 |
GO:1900740 |
regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.1 |
0.4 |
GO:0003221 |
right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.1 |
0.8 |
GO:0070562 |
regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 |
0.6 |
GO:0030242 |
pexophagy(GO:0030242) |
| 0.1 |
0.9 |
GO:2000124 |
regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 |
0.4 |
GO:1904742 |
protein poly-ADP-ribosylation(GO:0070212) regulation of telomeric DNA binding(GO:1904742) |
| 0.1 |
0.5 |
GO:0006570 |
tyrosine metabolic process(GO:0006570) ventricular zone neuroblast division(GO:0021847) |
| 0.1 |
1.3 |
GO:0035970 |
peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 |
0.3 |
GO:0060060 |
post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.1 |
0.4 |
GO:1900377 |
negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.1 |
0.4 |
GO:1904565 |
response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.1 |
0.3 |
GO:0015772 |
disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.1 |
0.7 |
GO:0060267 |
positive regulation of respiratory burst(GO:0060267) |
| 0.1 |
0.4 |
GO:0007171 |
activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 |
0.3 |
GO:0090271 |
positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.1 |
0.3 |
GO:0048818 |
embryonic nail plate morphogenesis(GO:0035880) positive regulation of hair follicle maturation(GO:0048818) positive regulation of catagen(GO:0051795) frontal suture morphogenesis(GO:0060364) |
| 0.1 |
0.5 |
GO:0036324 |
vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.1 |
0.2 |
GO:1902915 |
negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 |
0.5 |
GO:0021869 |
forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 |
1.3 |
GO:1902260 |
negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.1 |
0.3 |
GO:1902605 |
heterotrimeric G-protein complex assembly(GO:1902605) |
| 0.1 |
0.8 |
GO:0035095 |
behavioral response to nicotine(GO:0035095) |
| 0.1 |
0.4 |
GO:0033087 |
negative regulation of immature T cell proliferation(GO:0033087) |
| 0.1 |
0.5 |
GO:0072318 |
clathrin coat disassembly(GO:0072318) |
| 0.1 |
0.6 |
GO:0007195 |
adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.1 |
0.3 |
GO:0044830 |
modulation by host of viral RNA genome replication(GO:0044830) |
| 0.1 |
0.7 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 |
0.5 |
GO:0034587 |
piRNA metabolic process(GO:0034587) |
| 0.1 |
0.4 |
GO:0048245 |
eosinophil chemotaxis(GO:0048245) |
| 0.1 |
0.2 |
GO:0019085 |
early viral transcription(GO:0019085) |
| 0.1 |
0.2 |
GO:0042536 |
negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.1 |
0.2 |
GO:0042420 |
dopamine catabolic process(GO:0042420) |
| 0.1 |
0.6 |
GO:0006003 |
fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 |
0.4 |
GO:0032485 |
regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 |
0.8 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.1 |
0.1 |
GO:0097116 |
gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.1 |
1.4 |
GO:0001574 |
ganglioside biosynthetic process(GO:0001574) |
| 0.1 |
0.2 |
GO:0006933 |
negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.1 |
0.3 |
GO:0042997 |
negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.1 |
0.5 |
GO:0090073 |
positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 |
0.8 |
GO:0045974 |
miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 |
0.2 |
GO:2000481 |
positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 0.1 |
0.3 |
GO:0090148 |
membrane fission(GO:0090148) |
| 0.1 |
0.2 |
GO:0072362 |
regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.1 |
0.5 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 |
0.4 |
GO:0031848 |
protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 |
0.2 |
GO:0035063 |
nuclear speck organization(GO:0035063) |
| 0.1 |
0.2 |
GO:0002175 |
protein localization to paranode region of axon(GO:0002175) |
| 0.1 |
0.3 |
GO:0060527 |
prostate glandular acinus morphogenesis(GO:0060526) prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis(GO:0060527) |
| 0.1 |
0.5 |
GO:0060501 |
positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.1 |
0.1 |
GO:0035553 |
oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) oxidative demethylation(GO:0070989) |
| 0.1 |
0.3 |
GO:1900028 |
negative regulation of ruffle assembly(GO:1900028) |
| 0.1 |
0.5 |
GO:0051574 |
positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 |
0.2 |
GO:0050908 |
detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 |
0.3 |
GO:0035356 |
cellular triglyceride homeostasis(GO:0035356) |
| 0.1 |
0.2 |
GO:0061738 |
abscission(GO:0009838) late endosomal microautophagy(GO:0061738) positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.1 |
0.8 |
GO:0006120 |
mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 |
0.2 |
GO:0060023 |
soft palate development(GO:0060023) |
| 0.1 |
0.6 |
GO:0045217 |
cell-cell junction maintenance(GO:0045217) |
| 0.1 |
0.2 |
GO:0090526 |
regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 |
0.3 |
GO:0016198 |
axon choice point recognition(GO:0016198) |
| 0.0 |
0.4 |
GO:0032625 |
interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 |
0.8 |
GO:0060218 |
hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 |
0.2 |
GO:0032439 |
endosome localization(GO:0032439) |
| 0.0 |
0.1 |
GO:1904339 |
negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.0 |
0.2 |
GO:2000065 |
negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.0 |
0.1 |
GO:0044028 |
DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 |
0.2 |
GO:0007113 |
endomitotic cell cycle(GO:0007113) |
| 0.0 |
1.3 |
GO:0003301 |
physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 |
0.1 |
GO:0006667 |
sphinganine metabolic process(GO:0006667) |
| 0.0 |
0.1 |
GO:0001550 |
ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 |
0.1 |
GO:0048211 |
Golgi vesicle docking(GO:0048211) |
| 0.0 |
0.5 |
GO:0006107 |
oxaloacetate metabolic process(GO:0006107) |
| 0.0 |
0.4 |
GO:0034390 |
smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.0 |
0.1 |
GO:0035790 |
platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.0 |
0.1 |
GO:0046726 |
positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 |
0.1 |
GO:0045950 |
negative regulation of mitotic recombination(GO:0045950) |
| 0.0 |
0.1 |
GO:0071442 |
positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 |
0.1 |
GO:0015882 |
L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.0 |
0.4 |
GO:0014049 |
positive regulation of glutamate secretion(GO:0014049) |
| 0.0 |
0.4 |
GO:0061302 |
smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 |
0.1 |
GO:0002765 |
immune response-inhibiting signal transduction(GO:0002765) |
| 0.0 |
0.2 |
GO:0014816 |
skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 |
0.1 |
GO:0002329 |
pre-B cell differentiation(GO:0002329) |
| 0.0 |
0.5 |
GO:0007530 |
sex determination(GO:0007530) |
| 0.0 |
0.2 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.0 |
0.5 |
GO:0007216 |
G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 |
0.9 |
GO:0010842 |
retina layer formation(GO:0010842) |
| 0.0 |
1.0 |
GO:0060074 |
synapse maturation(GO:0060074) |
| 0.0 |
0.8 |
GO:0010880 |
regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) |
| 0.0 |
1.6 |
GO:0051489 |
regulation of filopodium assembly(GO:0051489) |
| 0.0 |
0.2 |
GO:0009235 |
cobalamin metabolic process(GO:0009235) |
| 0.0 |
0.1 |
GO:0034454 |
microtubule anchoring at centrosome(GO:0034454) |
| 0.0 |
1.4 |
GO:0008347 |
glial cell migration(GO:0008347) |
| 0.0 |
0.4 |
GO:2000651 |
positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 |
0.2 |
GO:0032782 |
bile acid secretion(GO:0032782) |
| 0.0 |
0.3 |
GO:0071578 |
zinc II ion transmembrane import(GO:0071578) |
| 0.0 |
0.2 |
GO:1900112 |
regulation of histone H3-K9 trimethylation(GO:1900112) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 |
0.1 |
GO:0050748 |
negative regulation of receptor biosynthetic process(GO:0010871) negative regulation of lipoprotein metabolic process(GO:0050748) |
| 0.0 |
0.1 |
GO:0035434 |
copper ion transmembrane transport(GO:0035434) |
| 0.0 |
0.1 |
GO:0007182 |
common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.0 |
0.1 |
GO:0051684 |
maintenance of Golgi location(GO:0051684) |
| 0.0 |
0.7 |
GO:0008045 |
motor neuron axon guidance(GO:0008045) |
| 0.0 |
0.2 |
GO:0006995 |
cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 |
0.3 |
GO:0071447 |
cellular response to hydroperoxide(GO:0071447) |
| 0.0 |
0.1 |
GO:0070886 |
positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 |
0.6 |
GO:0007608 |
sensory perception of smell(GO:0007608) |
| 0.0 |
0.2 |
GO:0070940 |
dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 |
0.4 |
GO:0080111 |
DNA demethylation(GO:0080111) |
| 0.0 |
0.1 |
GO:0043651 |
linoleic acid metabolic process(GO:0043651) |
| 0.0 |
0.2 |
GO:0018230 |
peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 |
0.3 |
GO:0042541 |
hemoglobin biosynthetic process(GO:0042541) |
| 0.0 |
0.1 |
GO:0042518 |
negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.0 |
0.4 |
GO:0008340 |
determination of adult lifespan(GO:0008340) |
| 0.0 |
0.1 |
GO:2000623 |
regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 |
0.3 |
GO:0043984 |
histone H4-K16 acetylation(GO:0043984) |
| 0.0 |
0.7 |
GO:0045494 |
photoreceptor cell maintenance(GO:0045494) |
| 0.0 |
0.1 |
GO:0072344 |
rescue of stalled ribosome(GO:0072344) |
| 0.0 |
0.1 |
GO:0046292 |
formaldehyde metabolic process(GO:0046292) |
| 0.0 |
0.1 |
GO:0006172 |
ADP biosynthetic process(GO:0006172) |
| 0.0 |
0.4 |
GO:0001833 |
inner cell mass cell proliferation(GO:0001833) |
| 0.0 |
0.8 |
GO:0060612 |
adipose tissue development(GO:0060612) |
| 0.0 |
0.3 |
GO:0010225 |
response to UV-C(GO:0010225) |
| 0.0 |
0.1 |
GO:0036092 |
phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 |
0.1 |
GO:0090274 |
positive regulation of somatostatin secretion(GO:0090274) |
| 0.0 |
0.3 |
GO:0030325 |
adrenal gland development(GO:0030325) |
| 0.0 |
0.1 |
GO:0070317 |
negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 |
0.3 |
GO:0070498 |
interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 |
0.1 |
GO:1903624 |
thymocyte apoptotic process(GO:0070242) regulation of apoptotic DNA fragmentation(GO:1902510) regulation of DNA catabolic process(GO:1903624) |
| 0.0 |
1.3 |
GO:0030032 |
lamellipodium assembly(GO:0030032) |
| 0.0 |
0.0 |
GO:0035754 |
B cell chemotaxis(GO:0035754) |
| 0.0 |
0.4 |
GO:0001783 |
B cell apoptotic process(GO:0001783) |
| 0.0 |
0.2 |
GO:0030322 |
stabilization of membrane potential(GO:0030322) |
| 0.0 |
0.1 |
GO:0035428 |
hexose transmembrane transport(GO:0035428) |
| 0.0 |
0.0 |
GO:2000508 |
regulation of dendritic cell chemotaxis(GO:2000508) positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.0 |
0.1 |
GO:0043922 |
negative regulation by host of viral transcription(GO:0043922) |
| 0.0 |
0.4 |
GO:0042773 |
ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 |
0.3 |
GO:0021702 |
cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 |
1.5 |
GO:0019722 |
calcium-mediated signaling(GO:0019722) |
| 0.0 |
0.3 |
GO:0003351 |
epithelial cilium movement(GO:0003351) |
| 0.0 |
0.7 |
GO:0045599 |
negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 |
0.0 |
GO:0060455 |
negative regulation of urine volume(GO:0035811) negative regulation of gastric acid secretion(GO:0060455) |
| 0.0 |
0.4 |
GO:0006101 |
tricarboxylic acid cycle(GO:0006099) citrate metabolic process(GO:0006101) |
| 0.0 |
0.5 |
GO:0043407 |
negative regulation of MAP kinase activity(GO:0043407) |
| 0.0 |
0.4 |
GO:0001569 |
patterning of blood vessels(GO:0001569) |
| 0.0 |
1.5 |
GO:0048706 |
embryonic skeletal system development(GO:0048706) |
| 0.0 |
0.5 |
GO:0000387 |
spliceosomal snRNP assembly(GO:0000387) |
| 0.0 |
0.2 |
GO:0002053 |
positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.0 |
0.1 |
GO:0007262 |
STAT protein import into nucleus(GO:0007262) |
| 0.0 |
0.1 |
GO:0060134 |
prepulse inhibition(GO:0060134) |
| 0.0 |
0.1 |
GO:0035520 |
monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 |
0.1 |
GO:1902965 |
protein localization to early endosome(GO:1902946) regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.0 |
0.1 |
GO:0031017 |
exocrine pancreas development(GO:0031017) |
| 0.0 |
0.3 |
GO:0071539 |
protein localization to centrosome(GO:0071539) |
| 0.0 |
0.1 |
GO:0070072 |
vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 |
0.5 |
GO:0046627 |
negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 |
0.2 |
GO:0038092 |
nodal signaling pathway(GO:0038092) |
| 0.0 |
0.2 |
GO:2000114 |
regulation of establishment of cell polarity(GO:2000114) |
| 0.0 |
0.0 |
GO:1900747 |
negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 |
0.0 |
GO:0007217 |
tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 |
0.1 |
GO:0007379 |
segment specification(GO:0007379) |
| 0.0 |
0.1 |
GO:0006449 |
regulation of translational termination(GO:0006449) |
| 0.0 |
0.1 |
GO:0007320 |
insemination(GO:0007320) |
| 0.0 |
1.2 |
GO:0007266 |
Rho protein signal transduction(GO:0007266) |
| 0.0 |
0.1 |
GO:0042753 |
positive regulation of circadian rhythm(GO:0042753) |
| 0.0 |
0.1 |
GO:0032570 |
response to progesterone(GO:0032570) |