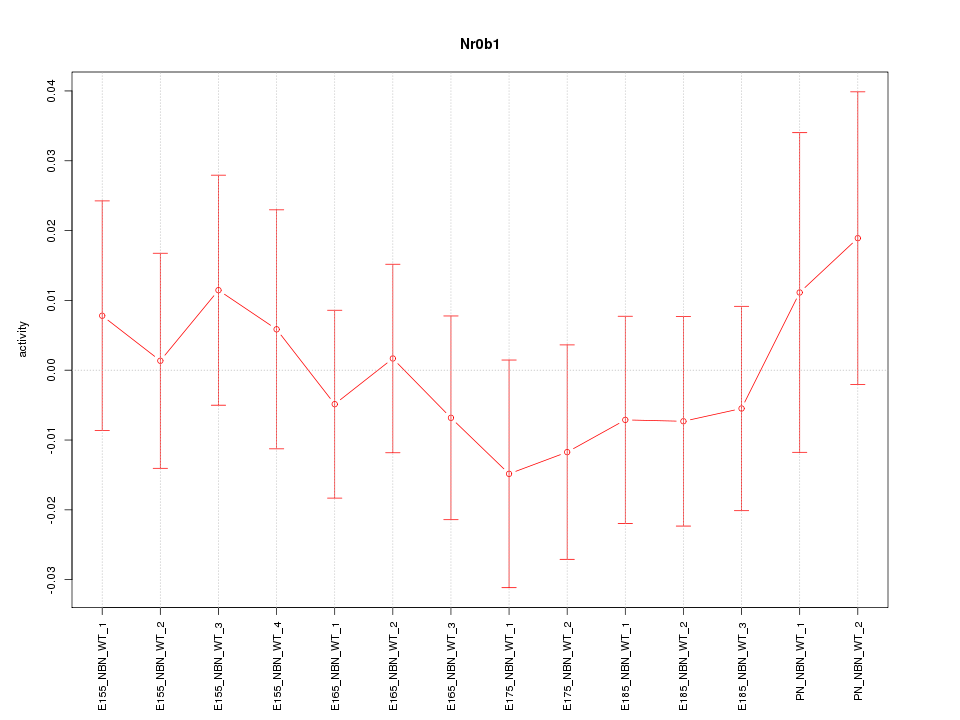
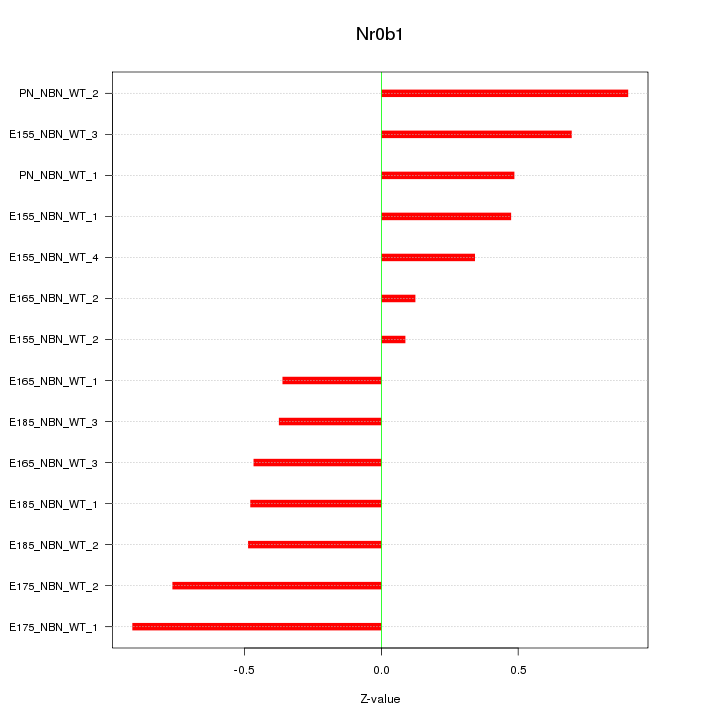
Motif ID: Nr0b1
Z-value: 0.552
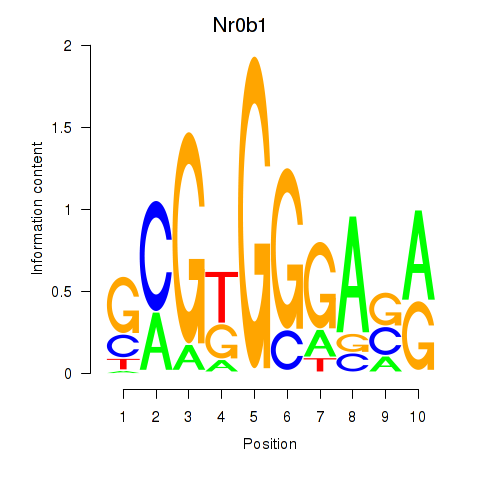
Transcription factors associated with Nr0b1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nr0b1 | ENSMUSG00000025056.4 | Nr0b1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr0b1 | mm10_v2_chrX_+_86191764_86191782 | 0.24 | 4.2e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.7 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 | 0.5 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.1 | 0.5 | GO:0007223 | muscular septum morphogenesis(GO:0003150) Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.1 | 0.3 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.1 | 0.3 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 0.2 | GO:2001045 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.3 | GO:0045759 | negative regulation of action potential(GO:0045759) |
| 0.1 | 0.2 | GO:0036166 | phenotypic switching(GO:0036166) |
| 0.1 | 0.2 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 | 0.7 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.0 | 0.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.6 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.1 | GO:0072070 | loop of Henle development(GO:0072070) metanephric loop of Henle development(GO:0072236) |
| 0.0 | 0.2 | GO:0051660 | cortical microtubule organization(GO:0043622) establishment of centrosome localization(GO:0051660) maintenance of centrosome location(GO:0051661) regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.4 | GO:0014842 | regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.0 | 0.1 | GO:1902915 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.2 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.6 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.0 | 0.2 | GO:0036289 | peptidyl-serine autophosphorylation(GO:0036289) |
| 0.0 | 0.3 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.3 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.2 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.0 | GO:0010643 | cell communication by chemical coupling(GO:0010643) negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.0 | 0.1 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 0.3 | GO:0002111 | BRCA2-BRAF35 complex(GO:0002111) |
| 0.1 | 0.9 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.7 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.2 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.6 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 1.1 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.0 | GO:1902737 | dendritic filopodium(GO:1902737) |
| 0.0 | 0.6 | GO:0045171 | intercellular bridge(GO:0045171) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 0.2 | GO:0071862 | protein phosphatase type 1 activator activity(GO:0071862) |
| 0.1 | 0.2 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 0.4 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.2 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 0.8 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 0.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.2 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.2 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.5 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.3 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |