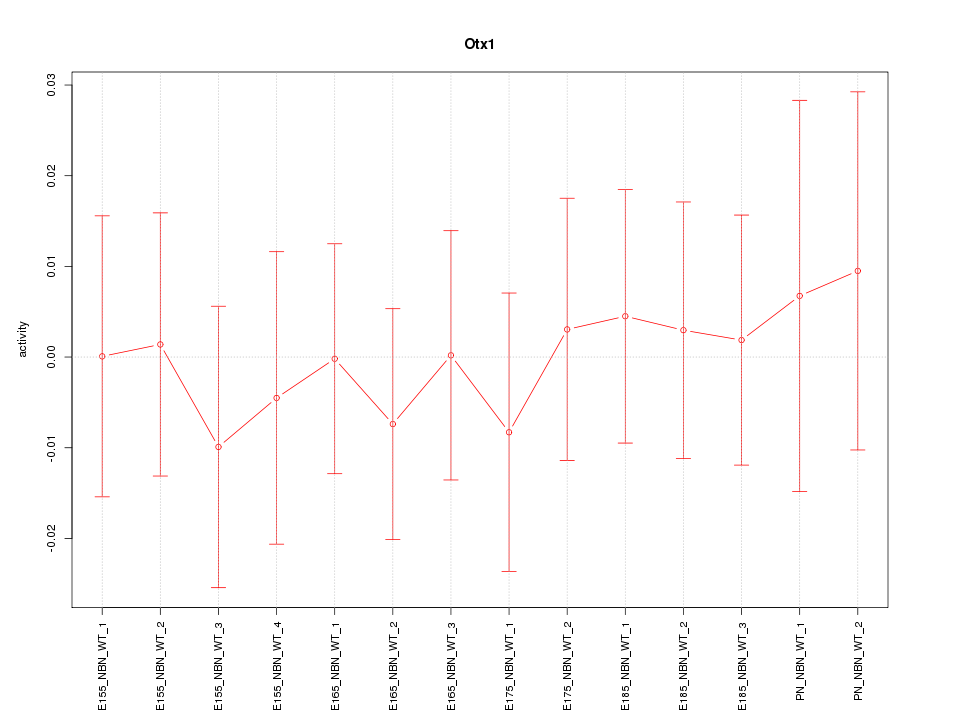
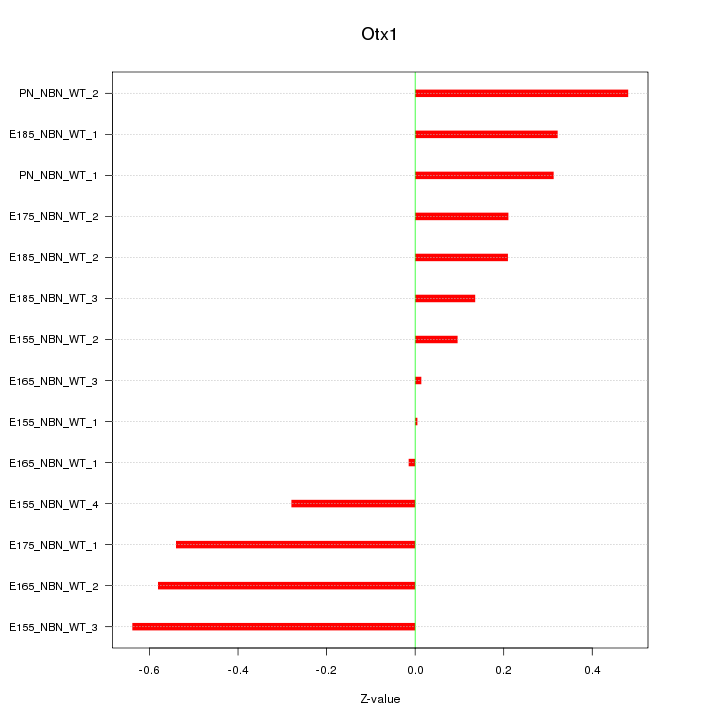
Motif ID: Otx1
Z-value: 0.345
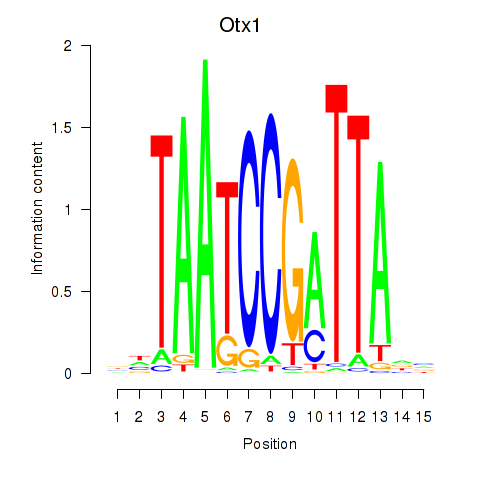
Transcription factors associated with Otx1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Otx1 | ENSMUSG00000005917.9 | Otx1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Otx1 | mm10_v2_chr11_-_22001605_22001631 | -0.45 | 1.1e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0002295 | T-helper cell lineage commitment(GO:0002295) interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.1 | 0.3 | GO:0097461 | ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) |
| 0.1 | 0.4 | GO:0072539 | T-helper 17 cell differentiation(GO:0072539) |
| 0.1 | 0.3 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.1 | 0.3 | GO:0097503 | sialylation(GO:0097503) |
| 0.1 | 0.5 | GO:0071474 | cellular hyperosmotic response(GO:0071474) |
| 0.1 | 0.2 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.3 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.4 | GO:0035814 | negative regulation of renal sodium excretion(GO:0035814) |
| 0.0 | 0.2 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 | 0.1 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 | 0.3 | GO:0051764 | actin crosslink formation(GO:0051764) positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.1 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.0 | 0.7 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.2 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 | 0.1 | GO:2001045 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.1 | GO:0032911 | nerve growth factor production(GO:0032902) negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 | 0.1 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.0 | 0.2 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.0 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.0 | 0.2 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.1 | 0.3 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.1 | 0.3 | GO:0043256 | laminin complex(GO:0043256) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.3 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.2 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.1 | 0.2 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.0 | 0.1 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.3 | GO:0008932 | lytic endotransglycosylase activity(GO:0008932) |
| 0.0 | 0.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0071862 | protein phosphatase type 1 activator activity(GO:0071862) |
| 0.0 | 0.8 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.1 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.0 | 0.0 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |