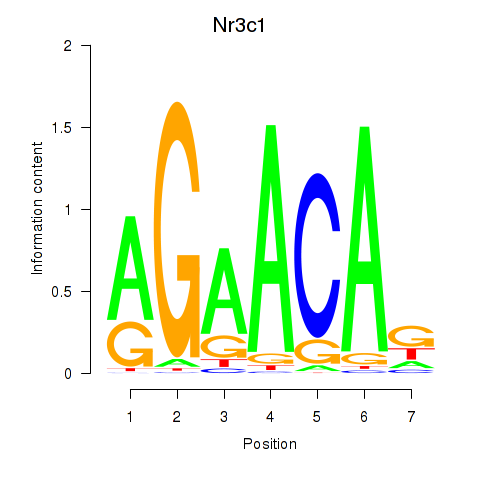
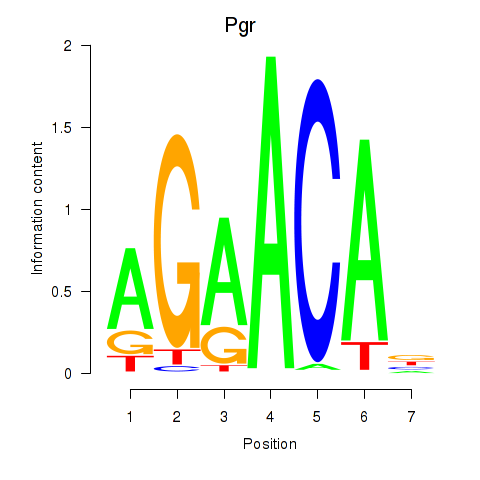
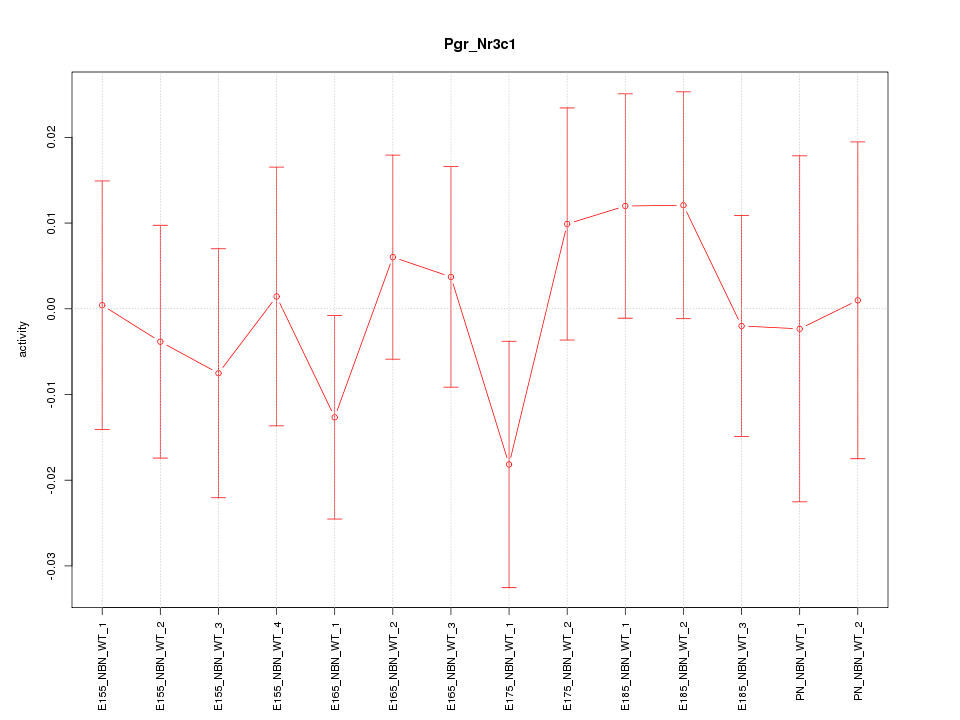
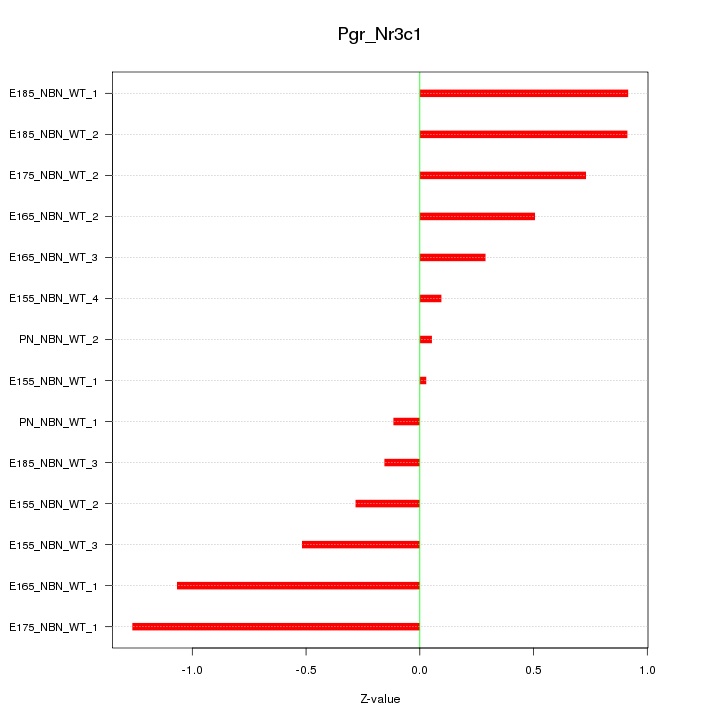
| 1.3 |
6.3 |
GO:0070327 |
thyroid hormone transport(GO:0070327) |
| 0.5 |
2.0 |
GO:0032914 |
positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.3 |
0.9 |
GO:0015889 |
cobalamin transport(GO:0015889) |
| 0.2 |
0.7 |
GO:0003100 |
regulation of systemic arterial blood pressure by endothelin(GO:0003100) regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.1 |
0.7 |
GO:0032534 |
regulation of microvillus assembly(GO:0032534) |
| 0.1 |
0.3 |
GO:0046671 |
negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.1 |
0.3 |
GO:2000383 |
regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.1 |
0.9 |
GO:2000653 |
regulation of genetic imprinting(GO:2000653) |
| 0.1 |
0.2 |
GO:0006601 |
creatine biosynthetic process(GO:0006601) |
| 0.1 |
0.4 |
GO:0051461 |
protein import into peroxisome matrix, docking(GO:0016560) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.1 |
0.6 |
GO:0010572 |
positive regulation of platelet activation(GO:0010572) |
| 0.1 |
1.1 |
GO:0006958 |
complement activation, classical pathway(GO:0006958) |
| 0.1 |
0.1 |
GO:1904425 |
negative regulation of GTP binding(GO:1904425) |
| 0.0 |
0.2 |
GO:0086013 |
membrane repolarization during cardiac muscle cell action potential(GO:0086013) |
| 0.0 |
0.1 |
GO:1902336 |
neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) positive regulation of retinal ganglion cell axon guidance(GO:1902336) |
| 0.0 |
0.3 |
GO:0015670 |
carbon dioxide transport(GO:0015670) |
| 0.0 |
0.1 |
GO:1903031 |
regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 |
0.3 |
GO:0050716 |
positive regulation of interleukin-1 secretion(GO:0050716) positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.0 |
0.2 |
GO:0015868 |
purine ribonucleotide transport(GO:0015868) |
| 0.0 |
0.1 |
GO:0046104 |
thymidine metabolic process(GO:0046104) |
| 0.0 |
0.2 |
GO:0060353 |
regulation of cell adhesion molecule production(GO:0060353) positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 |
0.1 |
GO:0008582 |
regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.0 |
0.2 |
GO:0048625 |
myoblast fate commitment(GO:0048625) |
| 0.0 |
0.1 |
GO:0003406 |
retinal pigment epithelium development(GO:0003406) |
| 0.0 |
0.1 |
GO:0010519 |
negative regulation of phospholipase activity(GO:0010519) |
| 0.0 |
0.0 |
GO:0071475 |
cellular hyperosmotic salinity response(GO:0071475) |