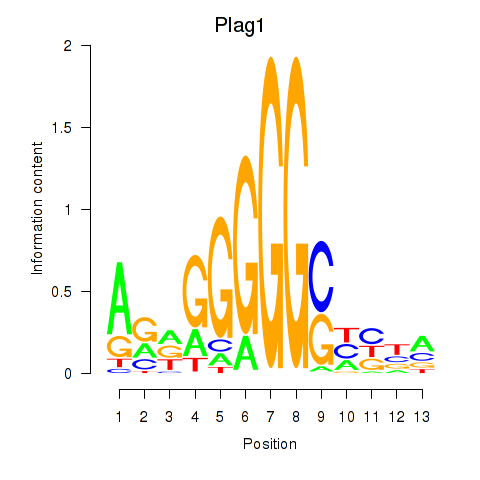
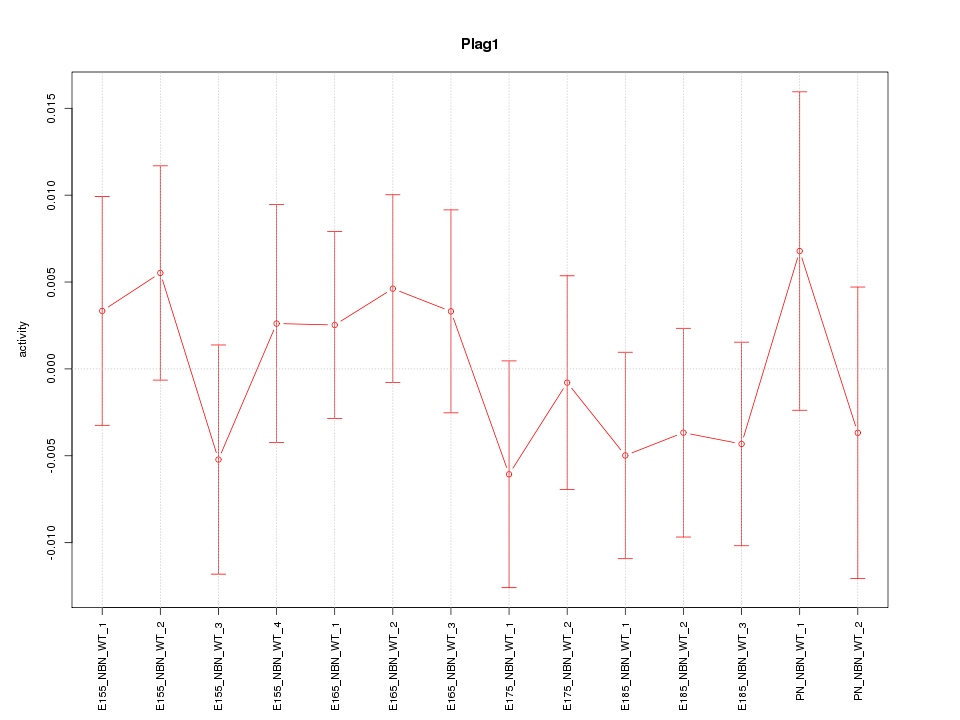
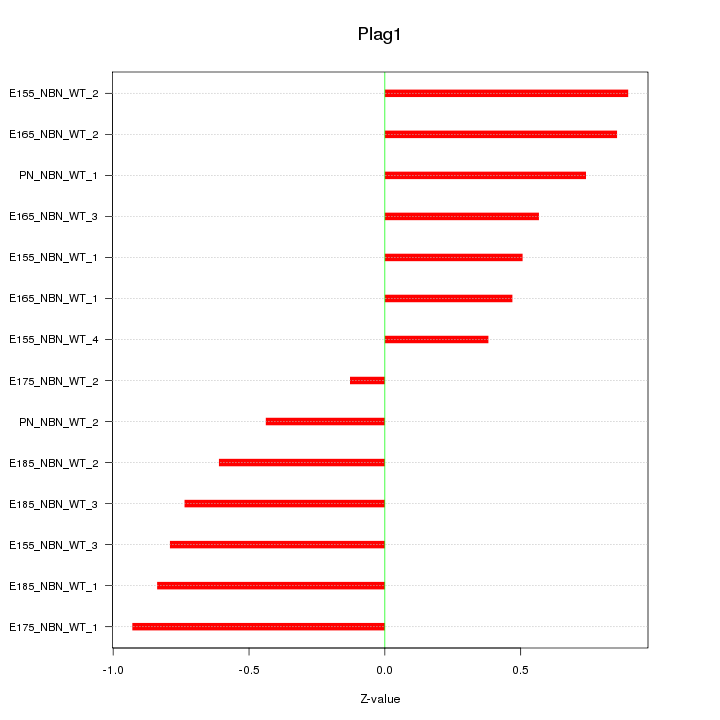
| 0.3 |
0.9 |
GO:0051902 |
negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.2 |
1.4 |
GO:2001199 |
negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.2 |
0.5 |
GO:0003195 |
tricuspid valve development(GO:0003175) tricuspid valve morphogenesis(GO:0003186) tricuspid valve formation(GO:0003195) right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.2 |
0.7 |
GO:0003150 |
muscular septum morphogenesis(GO:0003150) |
| 0.2 |
0.8 |
GO:0006680 |
glucosylceramide catabolic process(GO:0006680) negative regulation of filopodium assembly(GO:0051490) |
| 0.1 |
0.7 |
GO:0061642 |
chemoattraction of axon(GO:0061642) |
| 0.1 |
0.4 |
GO:0061144 |
alveolar secondary septum development(GO:0061144) |
| 0.1 |
0.4 |
GO:1904058 |
positive regulation of sensory perception of pain(GO:1904058) |
| 0.1 |
1.1 |
GO:0035542 |
regulation of SNARE complex assembly(GO:0035542) |
| 0.1 |
0.4 |
GO:1903225 |
negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 |
0.5 |
GO:0060022 |
hard palate development(GO:0060022) |
| 0.1 |
0.5 |
GO:1901204 |
positive regulation of guanylate cyclase activity(GO:0031284) regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 0.1 |
0.7 |
GO:0048852 |
diencephalon morphogenesis(GO:0048852) |
| 0.1 |
0.2 |
GO:0007223 |
Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.1 |
0.3 |
GO:1903244 |
positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.1 |
0.3 |
GO:0060598 |
dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.1 |
0.3 |
GO:0032053 |
ciliary basal body organization(GO:0032053) |
| 0.1 |
0.2 |
GO:0014738 |
regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.1 |
0.6 |
GO:0001842 |
neural fold formation(GO:0001842) |
| 0.1 |
0.2 |
GO:0070844 |
misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.1 |
0.4 |
GO:0045759 |
negative regulation of action potential(GO:0045759) |
| 0.1 |
0.4 |
GO:0061073 |
ciliary body morphogenesis(GO:0061073) |
| 0.1 |
0.3 |
GO:0033326 |
cerebrospinal fluid secretion(GO:0033326) |
| 0.1 |
0.5 |
GO:0033563 |
dorsal/ventral axon guidance(GO:0033563) |
| 0.1 |
0.2 |
GO:0007521 |
muscle cell fate determination(GO:0007521) |
| 0.1 |
0.2 |
GO:0021553 |
olfactory nerve development(GO:0021553) ureteric bud invasion(GO:0072092) |
| 0.1 |
0.2 |
GO:0090258 |
negative regulation of mitochondrial fission(GO:0090258) |
| 0.1 |
0.7 |
GO:0030322 |
stabilization of membrane potential(GO:0030322) |
| 0.1 |
0.2 |
GO:0015772 |
disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.1 |
1.3 |
GO:0090036 |
regulation of protein kinase C signaling(GO:0090036) |
| 0.1 |
0.2 |
GO:0016115 |
terpenoid catabolic process(GO:0016115) |
| 0.1 |
0.3 |
GO:0015803 |
branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 |
0.3 |
GO:0010528 |
regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.1 |
0.2 |
GO:0001830 |
trophectodermal cell fate commitment(GO:0001830) |
| 0.1 |
0.3 |
GO:0035022 |
positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.1 |
0.6 |
GO:0060272 |
embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.1 |
0.2 |
GO:0035604 |
fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) coronal suture morphogenesis(GO:0060365) squamous basal epithelial stem cell differentiation involved in prostate gland acinus development(GO:0060529) |
| 0.1 |
0.2 |
GO:0035984 |
response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.0 |
0.1 |
GO:0070889 |
platelet alpha granule organization(GO:0070889) |
| 0.0 |
0.2 |
GO:1902608 |
regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.0 |
0.1 |
GO:0003192 |
mitral valve formation(GO:0003192) |
| 0.0 |
0.1 |
GO:0048162 |
preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.0 |
0.3 |
GO:0045198 |
establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 |
0.1 |
GO:0003340 |
negative regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0003340) |
| 0.0 |
0.1 |
GO:1904431 |
positive regulation of t-circle formation(GO:1904431) |
| 0.0 |
0.1 |
GO:0035021 |
negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 |
0.3 |
GO:0072318 |
clathrin coat disassembly(GO:0072318) |
| 0.0 |
0.2 |
GO:0097411 |
regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 |
0.4 |
GO:0015816 |
glycine transport(GO:0015816) |
| 0.0 |
0.2 |
GO:1900748 |
positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 |
0.1 |
GO:0070269 |
pyroptosis(GO:0070269) |
| 0.0 |
0.1 |
GO:1903722 |
endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) late endosomal microautophagy(GO:0061738) regulation of centriole elongation(GO:1903722) |
| 0.0 |
0.3 |
GO:0061436 |
establishment of skin barrier(GO:0061436) |
| 0.0 |
1.5 |
GO:0007218 |
neuropeptide signaling pathway(GO:0007218) |
| 0.0 |
0.1 |
GO:0019482 |
beta-alanine metabolic process(GO:0019482) |
| 0.0 |
0.1 |
GO:0001869 |
regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.0 |
0.1 |
GO:1901675 |
negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 |
0.2 |
GO:0006384 |
transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 |
0.5 |
GO:0033234 |
negative regulation of protein sumoylation(GO:0033234) |
| 0.0 |
0.1 |
GO:1900145 |
regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900175) positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 |
0.3 |
GO:2000601 |
positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 |
0.2 |
GO:0042535 |
positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.0 |
0.3 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.0 |
0.3 |
GO:0002043 |
blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 |
0.0 |
GO:2000277 |
positive regulation of oxidative phosphorylation uncoupler activity(GO:2000277) |
| 0.0 |
0.1 |
GO:0032513 |
negative regulation of protein phosphatase type 2B activity(GO:0032513) |
| 0.0 |
0.1 |
GO:0002408 |
myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.0 |
0.3 |
GO:0038028 |
insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 |
0.1 |
GO:0021933 |
radial glia guided migration of cerebellar granule cell(GO:0021933) |
| 0.0 |
0.3 |
GO:0090051 |
negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.0 |
0.4 |
GO:0046697 |
decidualization(GO:0046697) |
| 0.0 |
0.1 |
GO:0055071 |
cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.0 |
0.0 |
GO:0014744 |
positive regulation of muscle adaptation(GO:0014744) |
| 0.0 |
0.3 |
GO:1900121 |
negative regulation of receptor binding(GO:1900121) |
| 0.0 |
0.0 |
GO:1903377 |
negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 |
0.1 |
GO:0043973 |
histone H3-K4 acetylation(GO:0043973) |
| 0.0 |
0.1 |
GO:0060264 |
regulation of respiratory burst involved in inflammatory response(GO:0060264) negative regulation of respiratory burst involved in inflammatory response(GO:0060266) negative regulation of respiratory burst(GO:0060268) |
| 0.0 |
0.2 |
GO:0034316 |
negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 |
0.2 |
GO:1903300 |
negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 |
0.1 |
GO:0050883 |
musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 |
0.1 |
GO:0071985 |
multivesicular body sorting pathway(GO:0071985) |
| 0.0 |
0.2 |
GO:0032464 |
positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 |
0.4 |
GO:0051988 |
regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 |
0.2 |
GO:0031848 |
protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 |
0.1 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.0 |
0.1 |
GO:0001543 |
ovarian follicle rupture(GO:0001543) |
| 0.0 |
0.1 |
GO:0097211 |
response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 |
0.7 |
GO:0030513 |
positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 |
0.2 |
GO:0097341 |
inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 |
0.1 |
GO:0043379 |
memory T cell differentiation(GO:0043379) |
| 0.0 |
0.1 |
GO:2000256 |
positive regulation of male germ cell proliferation(GO:2000256) |
| 0.0 |
0.1 |
GO:0071865 |
T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) positive regulation of T cell mediated immune response to tumor cell(GO:0002842) regulation of apoptotic process in bone marrow(GO:0071865) negative regulation of apoptotic process in bone marrow(GO:0071866) |
| 0.0 |
0.2 |
GO:0071763 |
nuclear membrane organization(GO:0071763) |
| 0.0 |
0.3 |
GO:0048096 |
chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 |
0.3 |
GO:0045671 |
negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 |
0.2 |
GO:0000320 |
re-entry into mitotic cell cycle(GO:0000320) |
| 0.0 |
0.2 |
GO:0001675 |
acrosome assembly(GO:0001675) |
| 0.0 |
0.0 |
GO:0010958 |
regulation of amino acid import(GO:0010958) regulation of L-arginine import(GO:0010963) |
| 0.0 |
0.7 |
GO:0060351 |
cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.0 |
0.1 |
GO:0070212 |
protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 |
0.3 |
GO:2000009 |
negative regulation of protein localization to cell surface(GO:2000009) |
| 0.0 |
0.2 |
GO:0051574 |
positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 |
0.1 |
GO:0071839 |
apoptotic process in bone marrow(GO:0071839) |
| 0.0 |
0.3 |
GO:0030574 |
collagen catabolic process(GO:0030574) |
| 0.0 |
0.4 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 0.0 |
0.1 |
GO:0042487 |
regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 |
0.2 |
GO:2000741 |
positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 |
0.2 |
GO:0042989 |
sequestering of actin monomers(GO:0042989) |
| 0.0 |
0.3 |
GO:0071378 |
growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 |
0.1 |
GO:0070966 |
nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 |
0.1 |
GO:0038128 |
ERBB2 signaling pathway(GO:0038128) |
| 0.0 |
0.1 |
GO:0034441 |
plasma lipoprotein particle oxidation(GO:0034441) |
| 0.0 |
0.2 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 |
0.3 |
GO:1902857 |
positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 |
0.1 |
GO:0046864 |
retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) alveolar primary septum development(GO:0061143) |
| 0.0 |
0.1 |
GO:0072610 |
interleukin-12 secretion(GO:0072610) regulation of interleukin-12 secretion(GO:2001182) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 |
0.1 |
GO:0021681 |
cerebellar granular layer development(GO:0021681) |
| 0.0 |
0.1 |
GO:0036353 |
histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 |
0.0 |
GO:0002765 |
immune response-inhibiting signal transduction(GO:0002765) |
| 0.0 |
0.1 |
GO:0006546 |
glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 |
0.1 |
GO:0006543 |
glutamine catabolic process(GO:0006543) |
| 0.0 |
0.1 |
GO:0032625 |
interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 |
0.1 |
GO:0003215 |
cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 |
0.2 |
GO:1902459 |
positive regulation of stem cell population maintenance(GO:1902459) |
| 0.0 |
0.1 |
GO:1903690 |
negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 |
0.1 |
GO:0000160 |
phosphorelay signal transduction system(GO:0000160) |
| 0.0 |
0.1 |
GO:0060040 |
retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 |
0.3 |
GO:0006474 |
N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 |
0.1 |
GO:2001197 |
regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 |
0.3 |
GO:0098743 |
cartilage condensation(GO:0001502) cell aggregation(GO:0098743) |
| 0.0 |
0.1 |
GO:0016561 |
protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.0 |
0.1 |
GO:0060373 |
regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 |
0.1 |
GO:0098734 |
protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 |
0.1 |
GO:0007320 |
insemination(GO:0007320) |
| 0.0 |
0.3 |
GO:0009409 |
response to cold(GO:0009409) |
| 0.0 |
0.1 |
GO:0006842 |
tricarboxylic acid transport(GO:0006842) succinate transport(GO:0015744) citrate transport(GO:0015746) |
| 0.0 |
0.1 |
GO:1900194 |
negative regulation of oocyte maturation(GO:1900194) |
| 0.0 |
0.2 |
GO:0035646 |
endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.0 |
0.1 |
GO:0015670 |
carbon dioxide transport(GO:0015670) |
| 0.0 |
0.2 |
GO:0070445 |
oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 |
0.1 |
GO:1902373 |
negative regulation of mRNA catabolic process(GO:1902373) |
| 0.0 |
0.4 |
GO:0016082 |
synaptic vesicle priming(GO:0016082) |
| 0.0 |
0.0 |
GO:0006667 |
sphinganine metabolic process(GO:0006667) |
| 0.0 |
0.1 |
GO:0060416 |
response to growth hormone(GO:0060416) |
| 0.0 |
0.0 |
GO:1901536 |
regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) |
| 0.0 |
0.2 |
GO:0071225 |
cellular response to muramyl dipeptide(GO:0071225) |
| 0.0 |
0.4 |
GO:0095500 |
acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 |
0.0 |
GO:0010643 |
cell communication by chemical coupling(GO:0010643) |
| 0.0 |
0.0 |
GO:0016344 |
meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.0 |
0.1 |
GO:1903416 |
negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) response to glycoside(GO:1903416) |
| 0.0 |
0.1 |
GO:0031053 |
primary miRNA processing(GO:0031053) |
| 0.0 |
0.1 |
GO:0051639 |
actin filament network formation(GO:0051639) |
| 0.0 |
0.1 |
GO:0006590 |
thyroid hormone generation(GO:0006590) |
| 0.0 |
0.0 |
GO:0019896 |
axonal transport of mitochondrion(GO:0019896) |
| 0.0 |
0.1 |
GO:0016259 |
selenocysteine metabolic process(GO:0016259) |
| 0.0 |
0.0 |
GO:0051934 |
positive regulation of synaptic transmission, dopaminergic(GO:0032226) positive regulation of neurotransmitter uptake(GO:0051582) dopamine uptake involved in synaptic transmission(GO:0051583) regulation of dopamine uptake involved in synaptic transmission(GO:0051584) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) catecholamine uptake involved in synaptic transmission(GO:0051934) regulation of catecholamine uptake involved in synaptic transmission(GO:0051940) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.0 |
0.0 |
GO:0070649 |
formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 |
0.2 |
GO:2000193 |
positive regulation of fatty acid transport(GO:2000193) |
| 0.0 |
0.1 |
GO:2001032 |
regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.0 |
0.0 |
GO:0060527 |
prostate glandular acinus morphogenesis(GO:0060526) prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis(GO:0060527) |
| 0.0 |
0.1 |
GO:0071447 |
cellular response to hydroperoxide(GO:0071447) |
| 0.0 |
0.0 |
GO:0033615 |
mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 |
0.0 |
GO:0060672 |
epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 |
0.8 |
GO:0035914 |
skeletal muscle cell differentiation(GO:0035914) |
| 0.0 |
0.1 |
GO:0002051 |
osteoblast fate commitment(GO:0002051) |
| 0.0 |
0.1 |
GO:0006968 |
cellular defense response(GO:0006968) |
| 0.0 |
0.2 |
GO:0045742 |
positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.0 |
0.1 |
GO:0019886 |
antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 |
0.1 |
GO:0032825 |
positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.0 |
0.1 |
GO:0051461 |
corticotropin secretion(GO:0051458) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 |
0.1 |
GO:0007220 |
Notch receptor processing(GO:0007220) |
| 0.0 |
0.1 |
GO:0036506 |
maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 |
0.0 |
GO:2000834 |
androgen secretion(GO:0035935) testosterone secretion(GO:0035936) negative regulation of glucagon secretion(GO:0070093) regulation of androgen secretion(GO:2000834) positive regulation of androgen secretion(GO:2000836) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.0 |
0.0 |
GO:1903960 |
negative regulation of anion transmembrane transport(GO:1903960) |
| 0.0 |
0.1 |
GO:0060768 |
epithelial cell proliferation involved in prostate gland development(GO:0060767) regulation of epithelial cell proliferation involved in prostate gland development(GO:0060768) negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 |
0.2 |
GO:0080111 |
DNA demethylation(GO:0080111) |
| 0.0 |
0.1 |
GO:0050942 |
positive regulation of pigment cell differentiation(GO:0050942) |
| 0.0 |
0.1 |
GO:0048386 |
positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 |
0.1 |
GO:0006654 |
phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 |
0.0 |
GO:0019249 |
lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) |
| 0.0 |
0.1 |
GO:0014886 |
transition between slow and fast fiber(GO:0014886) |
| 0.0 |
0.0 |
GO:0048069 |
eye pigmentation(GO:0048069) |
| 0.0 |
0.0 |
GO:1902276 |
pancreatic amylase secretion(GO:0036395) regulation of pancreatic amylase secretion(GO:1902276) |
| 0.0 |
0.1 |
GO:0051697 |
protein delipidation(GO:0051697) |
| 0.0 |
0.1 |
GO:0045187 |
regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 |
0.4 |
GO:0090278 |
negative regulation of peptide secretion(GO:0002792) negative regulation of peptide hormone secretion(GO:0090278) |