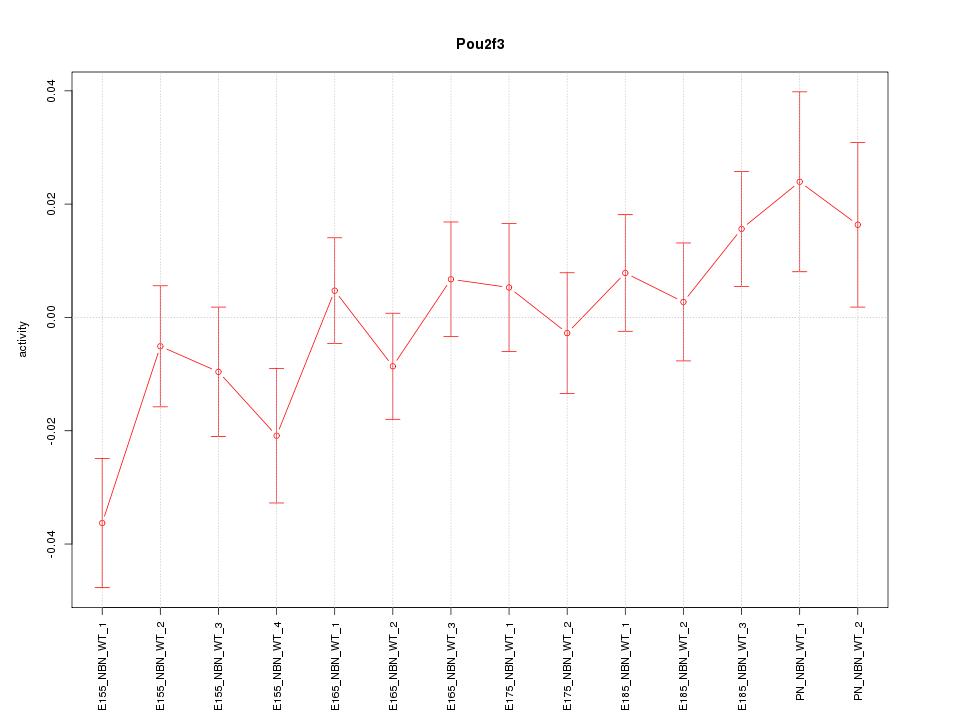
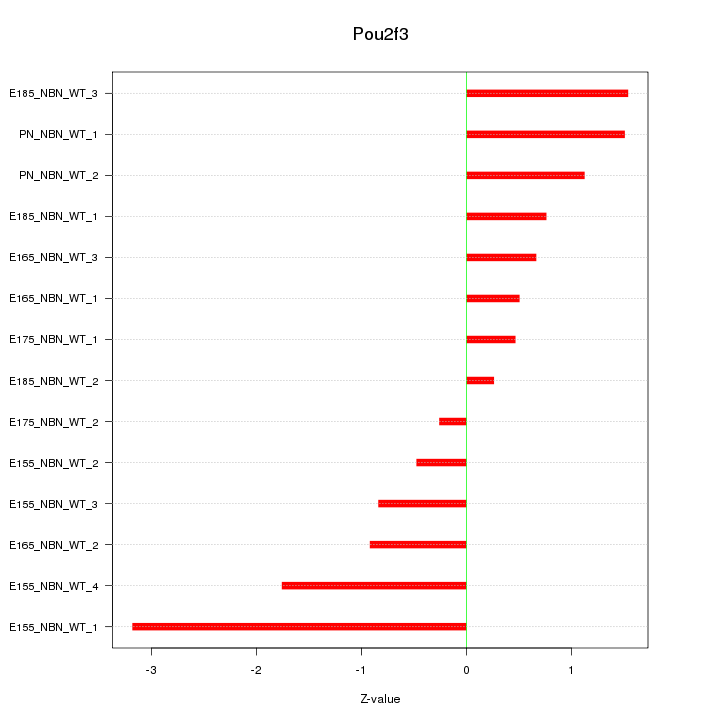
Motif ID: Pou2f3
Z-value: 1.269
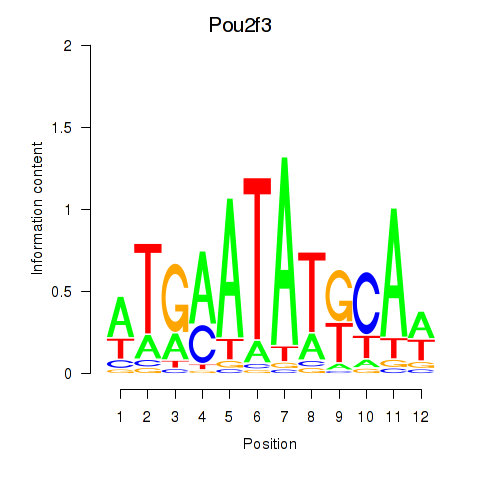
Transcription factors associated with Pou2f3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Pou2f3 | ENSMUSG00000032015.9 | Pou2f3 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:0008228 | opsonization(GO:0008228) modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.8 | 4.9 | GO:0035743 | CD4-positive, alpha-beta T cell cytokine production(GO:0035743) T-helper 2 cell cytokine production(GO:0035745) |
| 0.7 | 2.2 | GO:0019343 | cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) |
| 0.7 | 2.2 | GO:0002578 | negative regulation of antigen processing and presentation(GO:0002578) |
| 0.7 | 2.1 | GO:0051464 | positive regulation of cortisol secretion(GO:0051464) |
| 0.6 | 1.9 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.6 | 1.8 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.4 | 1.8 | GO:1903288 | regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.4 | 1.2 | GO:0030210 | heparin biosynthetic process(GO:0030210) Tie signaling pathway(GO:0048014) |
| 0.3 | 1.3 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.2 | 1.4 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.2 | 0.7 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.2 | 1.8 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.2 | 1.5 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.2 | 0.7 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.2 | 1.2 | GO:0003383 | apical constriction(GO:0003383) |
| 0.2 | 0.6 | GO:1902336 | neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) protein kinase D signaling(GO:0089700) positive regulation of retinal ganglion cell axon guidance(GO:1902336) |
| 0.2 | 1.1 | GO:0046909 | intermembrane transport(GO:0046909) |
| 0.2 | 1.0 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.2 | 0.9 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.7 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.1 | 0.7 | GO:0032621 | interleukin-18 production(GO:0032621) |
| 0.1 | 0.3 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 0.7 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.4 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.1 | 0.4 | GO:0009812 | flavonoid metabolic process(GO:0009812) flavonoid biosynthetic process(GO:0009813) flavonoid glucuronidation(GO:0052696) |
| 0.1 | 0.5 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 0.5 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 | 0.6 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.1 | 1.5 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 0.6 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.1 | 1.2 | GO:2000780 | negative regulation of double-strand break repair(GO:2000780) |
| 0.1 | 0.5 | GO:0002667 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.4 | GO:0071139 | resolution of recombination intermediates(GO:0071139) |
| 0.1 | 2.9 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 0.4 | GO:0072015 | ciliary body morphogenesis(GO:0061073) Notch signaling involved in heart development(GO:0061314) glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.1 | 0.4 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.1 | 2.3 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 0.6 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.1 | 0.8 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.1 | 0.8 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 0.3 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.1 | 0.9 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 0.6 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.1 | 0.5 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.0 | 1.3 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 1.5 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 0.3 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.2 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.2 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.2 | GO:0090037 | positive regulation of protein kinase C signaling(GO:0090037) |
| 0.0 | 1.3 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.2 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.1 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.2 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 1.3 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 0.8 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 1.0 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 0.4 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 2.6 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.1 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.0 | 0.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.5 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.1 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.2 | GO:0033147 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.3 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.0 | 0.1 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.2 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.8 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.0 | 0.2 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 1.0 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 0.2 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 1.3 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.0 | 0.3 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.8 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.2 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.2 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.2 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) negative regulation of phagocytosis(GO:0050765) |
| 0.0 | 0.1 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.4 | 2.1 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.3 | 1.3 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.3 | 1.3 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.3 | 1.3 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.3 | 1.8 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.2 | 0.7 | GO:0071920 | cleavage body(GO:0071920) |
| 0.1 | 0.3 | GO:0030894 | replisome(GO:0030894) |
| 0.1 | 0.7 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.1 | 1.0 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 0.5 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 0.5 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.8 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 1.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 0.5 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 0.9 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 1.5 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 0.2 | GO:0005940 | septin ring(GO:0005940) |
| 0.0 | 0.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.3 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 2.6 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.2 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.4 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 1.0 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 1.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 1.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.8 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 5.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 1.2 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.9 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 2.8 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.2 | GO:0031011 | Ino80 complex(GO:0031011) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.7 | 3.6 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.7 | 2.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.6 | 1.9 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.4 | 1.3 | GO:0043912 | D-lysine oxidase activity(GO:0043912) |
| 0.3 | 1.0 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.3 | 1.3 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.2 | 1.8 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.2 | 0.6 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.2 | 0.7 | GO:0103116 | alpha-D-galactofuranose transporter activity(GO:0103116) |
| 0.2 | 1.8 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.1 | 0.6 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 0.4 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.6 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.1 | 1.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 0.7 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.1 | 0.4 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.1 | 1.4 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 2.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.6 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.1 | 1.8 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.1 | 0.4 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 1.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.4 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 1.8 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 1.1 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 0.5 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 1.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 1.2 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.3 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 2.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.3 | GO:0052630 | CTP:2,3-di-O-geranylgeranyl-sn-glycero-1-phosphate cytidyltransferase activity(GO:0043338) phospholactate guanylyltransferase activity(GO:0043814) ATP:coenzyme F420 adenylyltransferase activity(GO:0043910) UDP-N-acetylgalactosamine diphosphorylase activity(GO:0052630) |
| 0.0 | 0.6 | GO:0070636 | single-strand selective uracil DNA N-glycosylase activity(GO:0017065) nicotinamide riboside hydrolase activity(GO:0070635) nicotinic acid riboside hydrolase activity(GO:0070636) deoxyribonucleoside 5'-monophosphate N-glycosidase activity(GO:0070694) |
| 0.0 | 0.4 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.1 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 0.7 | GO:0043747 | protein-N-terminal asparagine amidohydrolase activity(GO:0008418) UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase activity(GO:0008759) iprodione amidohydrolase activity(GO:0018748) (3,5-dichlorophenylurea)acetate amidohydrolase activity(GO:0018749) 4'-(2-hydroxyisopropyl)phenylurea amidohydrolase activity(GO:0034571) didemethylisoproturon amidohydrolase activity(GO:0034573) N-isopropylacetanilide amidohydrolase activity(GO:0034576) N-cyclohexylformamide amidohydrolase activity(GO:0034781) isonicotinic acid hydrazide hydrolase activity(GO:0034876) cis-aconitamide amidase activity(GO:0034882) gamma-N-formylaminovinylacetate hydrolase activity(GO:0034885) N2-acetyl-L-lysine deacetylase activity(GO:0043747) O-succinylbenzoate synthase activity(GO:0043748) indoleacetamide hydrolase activity(GO:0043864) N-acetylcitrulline deacetylase activity(GO:0043909) N-acetylgalactosamine-6-phosphate deacetylase activity(GO:0047419) diacetylchitobiose deacetylase activity(GO:0052773) chitooligosaccharide deacetylase activity(GO:0052790) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0086038 | calcium:sodium antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086038) |
| 0.0 | 0.8 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 3.3 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.2 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.5 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.4 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.2 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.5 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.5 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.5 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.0 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.7 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.8 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.8 | GO:0005080 | protein kinase C binding(GO:0005080) |