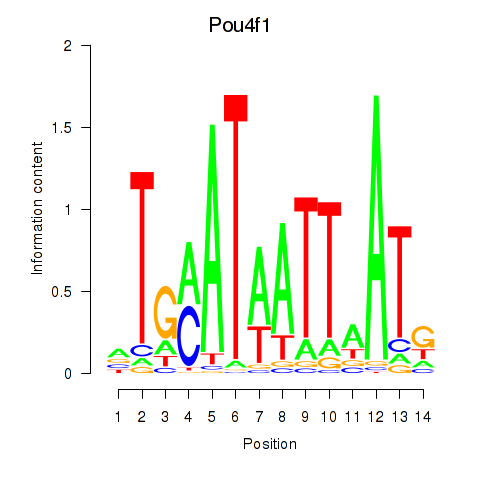
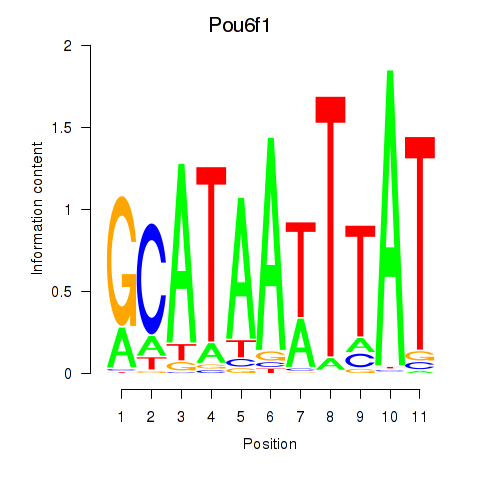
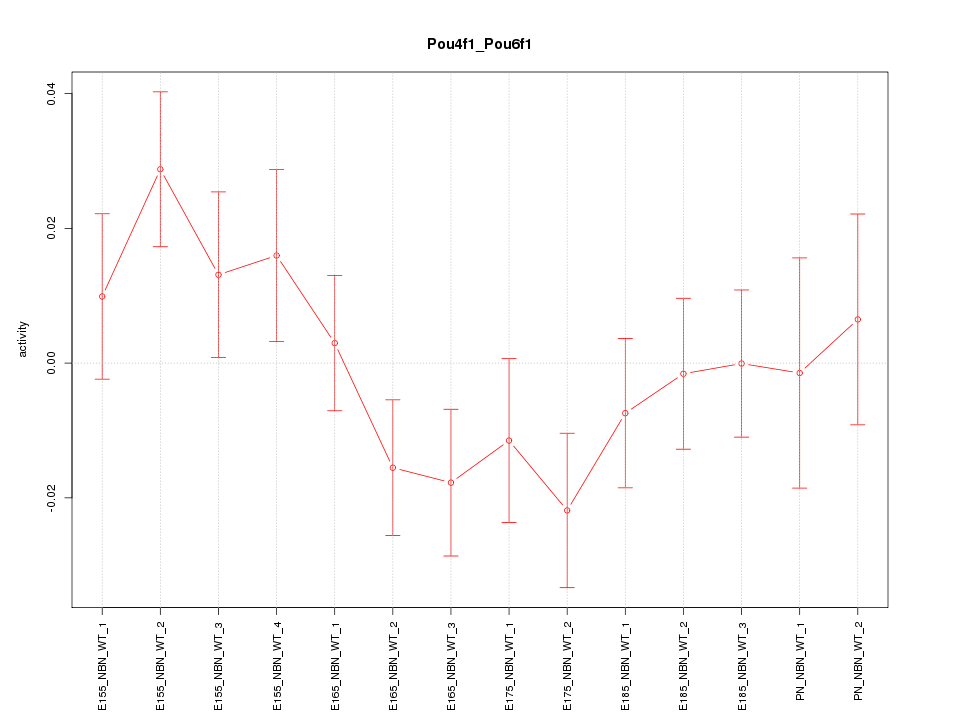
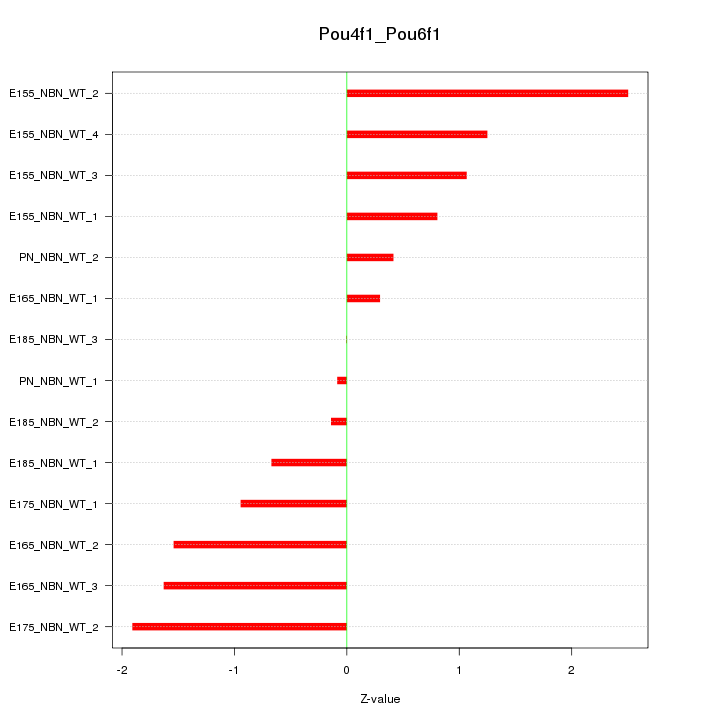
| 0.7 |
4.6 |
GO:0031536 |
positive regulation of exit from mitosis(GO:0031536) |
| 0.5 |
7.7 |
GO:1902993 |
positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.5 |
1.5 |
GO:0007621 |
negative regulation of female receptivity(GO:0007621) |
| 0.5 |
0.9 |
GO:0034372 |
very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.4 |
1.5 |
GO:0014846 |
esophagus smooth muscle contraction(GO:0014846) |
| 0.4 |
1.8 |
GO:0002457 |
T cell antigen processing and presentation(GO:0002457) |
| 0.3 |
1.0 |
GO:0071139 |
positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) resolution of recombination intermediates(GO:0071139) |
| 0.2 |
1.6 |
GO:0002681 |
somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.2 |
3.0 |
GO:0097090 |
presynaptic membrane organization(GO:0097090) |
| 0.1 |
1.2 |
GO:0010587 |
miRNA catabolic process(GO:0010587) |
| 0.1 |
0.6 |
GO:0036337 |
Fas signaling pathway(GO:0036337) |
| 0.1 |
0.8 |
GO:0097460 |
ferrous iron import into cell(GO:0097460) |
| 0.1 |
0.3 |
GO:0003221 |
right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.1 |
1.3 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 0.1 |
0.5 |
GO:0072257 |
metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.1 |
0.1 |
GO:0031394 |
positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 |
0.3 |
GO:0031860 |
telomeric 3' overhang formation(GO:0031860) |
| 0.1 |
0.9 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 |
0.4 |
GO:0071699 |
olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.1 |
0.8 |
GO:0060501 |
positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.1 |
0.2 |
GO:0009414 |
response to water deprivation(GO:0009414) |
| 0.1 |
1.0 |
GO:0001780 |
neutrophil homeostasis(GO:0001780) regulation of high voltage-gated calcium channel activity(GO:1901841) |
| 0.1 |
0.5 |
GO:0033147 |
negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.1 |
0.3 |
GO:0071787 |
endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 |
1.2 |
GO:0007097 |
nuclear migration(GO:0007097) |
| 0.1 |
0.2 |
GO:1904685 |
positive regulation of metalloendopeptidase activity(GO:1904685) |
| 0.1 |
0.2 |
GO:0035790 |
platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.1 |
0.3 |
GO:0021856 |
cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) trigeminal ganglion development(GO:0061551) facioacoustic ganglion development(GO:1903375) |
| 0.1 |
0.2 |
GO:1904211 |
membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.0 |
0.9 |
GO:0042541 |
hemoglobin biosynthetic process(GO:0042541) |
| 0.0 |
0.3 |
GO:0003406 |
retinal pigment epithelium development(GO:0003406) |
| 0.0 |
0.1 |
GO:0014877 |
response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 |
0.2 |
GO:0047484 |
regulation of response to osmotic stress(GO:0047484) |
| 0.0 |
0.4 |
GO:0048386 |
positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 |
0.2 |
GO:0010792 |
DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 |
0.2 |
GO:1903690 |
negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 |
2.2 |
GO:0045773 |
positive regulation of axon extension(GO:0045773) |
| 0.0 |
0.1 |
GO:0061743 |
motor learning(GO:0061743) |
| 0.0 |
0.3 |
GO:0007296 |
vitellogenesis(GO:0007296) |
| 0.0 |
0.3 |
GO:0043206 |
extracellular fibril organization(GO:0043206) |
| 0.0 |
0.4 |
GO:2000480 |
negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 |
0.1 |
GO:0048254 |
snoRNA localization(GO:0048254) |
| 0.0 |
0.2 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |
| 0.0 |
0.1 |
GO:0090258 |
negative regulation of mitochondrial fission(GO:0090258) |
| 0.0 |
0.2 |
GO:0090074 |
negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 |
0.4 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 |
0.2 |
GO:0031119 |
tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 |
0.3 |
GO:0060746 |
parental behavior(GO:0060746) |
| 0.0 |
0.3 |
GO:0043922 |
negative regulation by host of viral transcription(GO:0043922) |
| 0.0 |
0.3 |
GO:0046597 |
negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 |
0.1 |
GO:0042536 |
negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.0 |
0.2 |
GO:0000712 |
resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 |
0.1 |
GO:0089711 |
L-glutamate transmembrane transport(GO:0089711) |
| 0.0 |
0.1 |
GO:0044861 |
protein transport into plasma membrane raft(GO:0044861) |
| 0.0 |
0.1 |
GO:0061635 |
regulation of protein complex stability(GO:0061635) |
| 0.0 |
0.7 |
GO:0045494 |
photoreceptor cell maintenance(GO:0045494) |
| 0.0 |
0.5 |
GO:0035335 |
peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 |
0.1 |
GO:0072318 |
clathrin coat disassembly(GO:0072318) |
| 0.0 |
0.0 |
GO:0086046 |
membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 |
0.2 |
GO:1903608 |
protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 |
0.1 |
GO:0035428 |
hexose transmembrane transport(GO:0035428) |
| 0.0 |
0.0 |
GO:0051030 |
snRNA transport(GO:0051030) |
| 0.0 |
0.1 |
GO:0042572 |
retinol metabolic process(GO:0042572) |
| 0.0 |
0.2 |
GO:0060837 |
blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 |
0.2 |
GO:0021702 |
cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 |
0.7 |
GO:0010633 |
negative regulation of epithelial cell migration(GO:0010633) |
| 0.0 |
0.1 |
GO:0021942 |
radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 |
0.4 |
GO:0042773 |
ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 |
0.0 |
GO:0002331 |
pre-B cell allelic exclusion(GO:0002331) |
| 0.0 |
0.3 |
GO:0071385 |
cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 |
0.0 |
GO:0055071 |
cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.0 |
0.1 |
GO:0042136 |
neurotransmitter biosynthetic process(GO:0042136) |
| 0.0 |
0.8 |
GO:0043488 |
regulation of mRNA stability(GO:0043488) |
| 0.0 |
0.6 |
GO:1904893 |
negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 |
0.1 |
GO:0046548 |
retinal rod cell development(GO:0046548) |