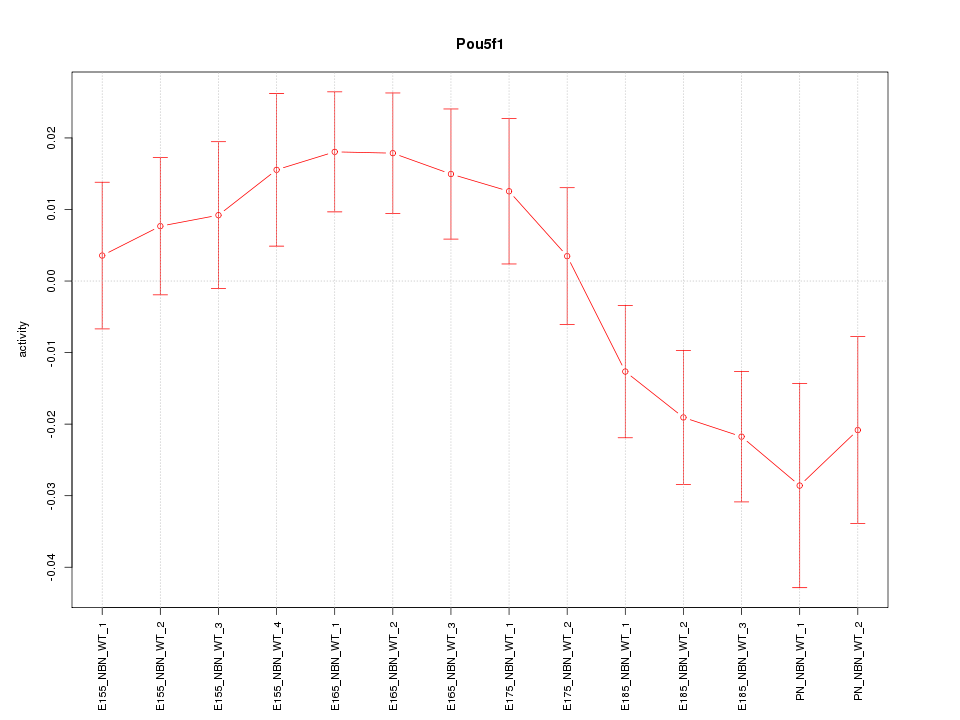
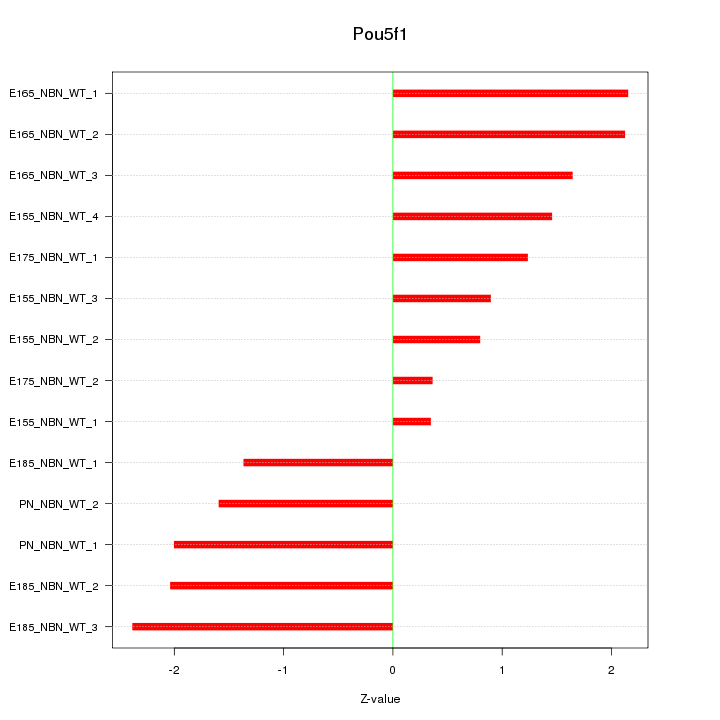
Motif ID: Pou5f1
Z-value: 1.591
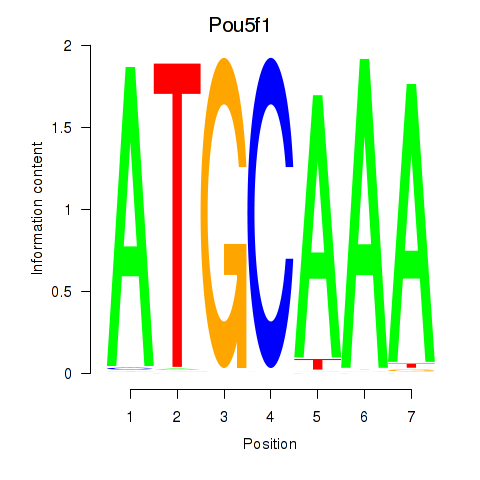
Transcription factors associated with Pou5f1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Pou5f1 | ENSMUSG00000024406.10 | Pou5f1 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 6.8 | GO:0055095 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 1.3 | 3.9 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 1.3 | 7.6 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 1.2 | 3.6 | GO:0060596 | mammary placode formation(GO:0060596) |
| 1.1 | 2.1 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.9 | 14.2 | GO:1902993 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.9 | 7.1 | GO:0032308 | positive regulation of prostaglandin secretion(GO:0032308) |
| 0.9 | 6.1 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.8 | 5.8 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.8 | 5.7 | GO:0034047 | regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.7 | 3.7 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.7 | 3.7 | GO:2000587 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.6 | 1.9 | GO:0030070 | insulin processing(GO:0030070) |
| 0.6 | 2.5 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.6 | 3.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.6 | 2.8 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.5 | 1.6 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.5 | 4.0 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.5 | 0.9 | GO:0030539 | male genitalia development(GO:0030539) |
| 0.4 | 5.1 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.4 | 1.2 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.4 | 2.0 | GO:0042636 | negative regulation of hair cycle(GO:0042636) progesterone secretion(GO:0042701) negative regulation of hair follicle development(GO:0051799) positive regulation of ovulation(GO:0060279) |
| 0.4 | 5.5 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.3 | 1.0 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.3 | 1.0 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.3 | 2.0 | GO:2001012 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.3 | 1.6 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.3 | 1.3 | GO:2000110 | protein sialylation(GO:1990743) negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.3 | 2.0 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.3 | 8.3 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.3 | 4.2 | GO:0006337 | nucleosome disassembly(GO:0006337) protein-DNA complex disassembly(GO:0032986) |
| 0.3 | 2.4 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.3 | 3.8 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.3 | 0.9 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.3 | 3.5 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.3 | 1.9 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.3 | 0.8 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.3 | 1.0 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.3 | 0.5 | GO:1901228 | positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.2 | 1.7 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.2 | 0.5 | GO:0061054 | dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) |
| 0.2 | 1.6 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.2 | 1.8 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.2 | 0.8 | GO:2000054 | regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.2 | 0.8 | GO:0032512 | regulation of protein phosphatase type 2B activity(GO:0032512) positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.2 | 1.0 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.2 | 3.2 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) |
| 0.2 | 0.8 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.2 | 0.5 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.2 | 0.7 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.2 | 1.3 | GO:0031053 | production of siRNA involved in RNA interference(GO:0030422) primary miRNA processing(GO:0031053) |
| 0.1 | 1.8 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 2.1 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 0.4 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.1 | 0.4 | GO:0097491 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.1 | 1.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.1 | 1.0 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.1 | 0.4 | GO:1902950 | regulation of dendritic spine maintenance(GO:1902950) positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.1 | 0.4 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.1 | 1.8 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 1.7 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 0.7 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.1 | 0.7 | GO:1903056 | regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.1 | 0.7 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.1 | 0.3 | GO:0010871 | negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.1 | 1.5 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.1 | 2.9 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.1 | 0.5 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 4.9 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 | 5.2 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.1 | 2.7 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 1.1 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.1 | 0.4 | GO:0001696 | gastric acid secretion(GO:0001696) |
| 0.1 | 1.4 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 1.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.8 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.1 | 1.8 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 1.1 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.1 | 1.1 | GO:0048305 | immunoglobulin secretion(GO:0048305) |
| 0.1 | 1.2 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.1 | 1.9 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 0.2 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 | 2.2 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.1 | 2.3 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 0.4 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.4 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 1.1 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 0.4 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.5 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.2 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.3 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.0 | 0.9 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.3 | GO:1900194 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 1.2 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 0.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 1.2 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 3.8 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 1.2 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 0.1 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 1.6 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.0 | 0.4 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.4 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 1.7 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 1.9 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.2 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 3.6 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.1 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.2 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.8 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.4 | GO:0001935 | endothelial cell proliferation(GO:0001935) |
| 0.0 | 0.9 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 2.2 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.0 | 0.3 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.0 | 2.0 | GO:0030307 | positive regulation of cell growth(GO:0030307) |
| 0.0 | 0.2 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.0 | 0.1 | GO:0038030 | non-canonical Wnt signaling pathway via MAPK cascade(GO:0038030) non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.4 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 2.1 | GO:0042384 | cilium assembly(GO:0042384) |
| 0.0 | 0.9 | GO:0000045 | autophagosome assembly(GO:0000045) |
| 0.0 | 0.0 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.9 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.3 | GO:0007623 | circadian rhythm(GO:0007623) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.6 | 1.8 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.5 | 7.7 | GO:0043196 | varicosity(GO:0043196) |
| 0.4 | 1.3 | GO:1903095 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.4 | 2.1 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.3 | 2.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.3 | 2.1 | GO:0042627 | chylomicron(GO:0042627) |
| 0.3 | 5.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 7.6 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.2 | 1.2 | GO:0044308 | axonal spine(GO:0044308) |
| 0.2 | 3.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.2 | 1.7 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.2 | 2.4 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.2 | 1.8 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.2 | 1.2 | GO:0000805 | X chromosome(GO:0000805) |
| 0.1 | 0.4 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.1 | 1.3 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 2.5 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 0.5 | GO:0002111 | BRCA2-BRAF35 complex(GO:0002111) |
| 0.1 | 4.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 3.5 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 1.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 1.7 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 0.4 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 5.7 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 7.2 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 0.4 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.9 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.1 | 2.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 0.9 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.7 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.9 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 9.9 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 2.2 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.8 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 1.6 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.5 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 1.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 2.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.4 | GO:0048188 | MLL3/4 complex(GO:0044666) Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 1.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 2.9 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 2.7 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.2 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.2 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 6.8 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.8 | 2.4 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 0.7 | 4.0 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.7 | 2.0 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.5 | 1.6 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.5 | 6.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.5 | 1.8 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.4 | 5.7 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.4 | 1.8 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.3 | 1.0 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.3 | 2.0 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.3 | 1.6 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.3 | 2.7 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.3 | 0.9 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 0.3 | 1.3 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.2 | 2.2 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.2 | 1.5 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.2 | 1.7 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.2 | 7.1 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.2 | 1.7 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.2 | 1.7 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.2 | 1.0 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.2 | 2.8 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 0.8 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.2 | 2.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.2 | 0.7 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.2 | 0.7 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.2 | 3.7 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.2 | 0.8 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 1.2 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.1 | 0.9 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 13.7 | GO:0019210 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.1 | 0.5 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.1 | 0.7 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.4 | GO:0048045 | 4-hydroxybenzoate octaprenyltransferase activity(GO:0008412) protoheme IX farnesyltransferase activity(GO:0008495) (S)-2,3-di-O-geranylgeranylglyceryl phosphate synthase activity(GO:0043888) cadaverine aminopropyltransferase activity(GO:0043918) agmatine aminopropyltransferase activity(GO:0043919) 1,4-dihydroxy-2-naphthoate octaprenyltransferase activity(GO:0046428) trans-pentaprenyltranstransferase activity(GO:0048045) ATP dimethylallyltransferase activity(GO:0052622) ADP dimethylallyltransferase activity(GO:0052623) |
| 0.1 | 3.8 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 1.1 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.1 | 2.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 1.3 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 0.9 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 3.8 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 2.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 2.5 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 0.4 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.1 | 0.9 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 1.7 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.3 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.4 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 0.5 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.8 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.9 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.0 | 0.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.8 | GO:0042556 | cobinamide kinase activity(GO:0008819) phytol kinase activity(GO:0010276) phenol kinase activity(GO:0018720) cyclin-dependent protein kinase activating kinase regulator activity(GO:0019914) inositol tetrakisphosphate 2-kinase activity(GO:0032942) heptose 7-phosphate kinase activity(GO:0033785) aminoglycoside phosphotransferase activity(GO:0034071) eukaryotic elongation factor-2 kinase regulator activity(GO:0042556) eukaryotic elongation factor-2 kinase activator activity(GO:0042557) LPPG:FO 2-phospho-L-lactate transferase activity(GO:0043743) cytidine kinase activity(GO:0043771) glycerate 2-kinase activity(GO:0043798) (S)-lactate 2-kinase activity(GO:0043841) phosphoserine:homoserine phosphotransferase activity(GO:0043899) L-seryl-tRNA(Sec) kinase activity(GO:0043915) phosphocholine transferase activity(GO:0044605) GTP-dependent polynucleotide kinase activity(GO:0051735) farnesol kinase activity(GO:0052668) CTP:2-trans,-6-trans-farnesol kinase activity(GO:0052669) geraniol kinase activity(GO:0052670) geranylgeraniol kinase activity(GO:0052671) CTP:geranylgeraniol kinase activity(GO:0052672) prenol kinase activity(GO:0052673) 1-phosphatidylinositol-5-kinase activity(GO:0052810) 1-phosphatidylinositol-3-phosphate 4-kinase activity(GO:0052811) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 1.8 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 1.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 2.8 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 3.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 4.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 2.0 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.6 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 1.8 | GO:0044824 | integrase activity(GO:0008907) T/G mismatch-specific endonuclease activity(GO:0043765) retroviral integrase activity(GO:0044823) retroviral 3' processing activity(GO:0044824) |
| 0.0 | 0.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.3 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.0 | 1.4 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0001565 | phorbol ester receptor activity(GO:0001565) non-kinase phorbol ester receptor activity(GO:0001566) |
| 0.0 | 0.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.5 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.4 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 1.8 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.3 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 1.2 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.8 | GO:0018602 | sulfonate dioxygenase activity(GO:0000907) 2,4-dichlorophenoxyacetate alpha-ketoglutarate dioxygenase activity(GO:0018602) hypophosphite dioxygenase activity(GO:0034792) gibberellin 2-beta-dioxygenase activity(GO:0045543) C-19 gibberellin 2-beta-dioxygenase activity(GO:0052634) C-20 gibberellin 2-beta-dioxygenase activity(GO:0052635) |
| 0.0 | 0.1 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 0.6 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 1.4 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 2.3 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 1.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 5.2 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.3 | GO:0032183 | SUMO binding(GO:0032183) |