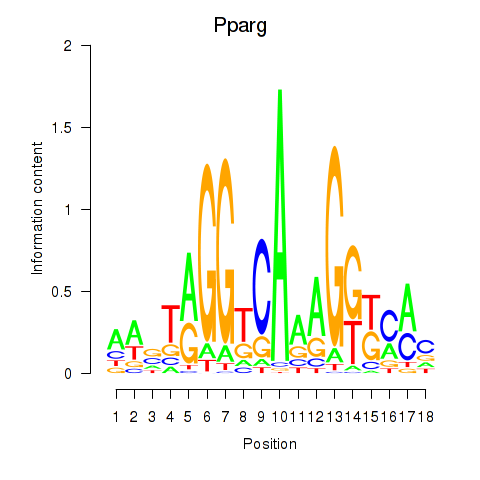
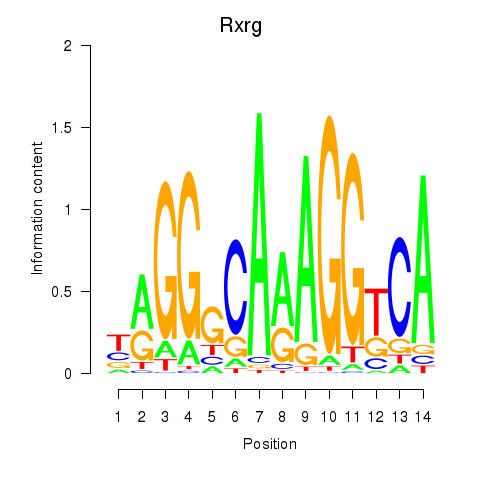
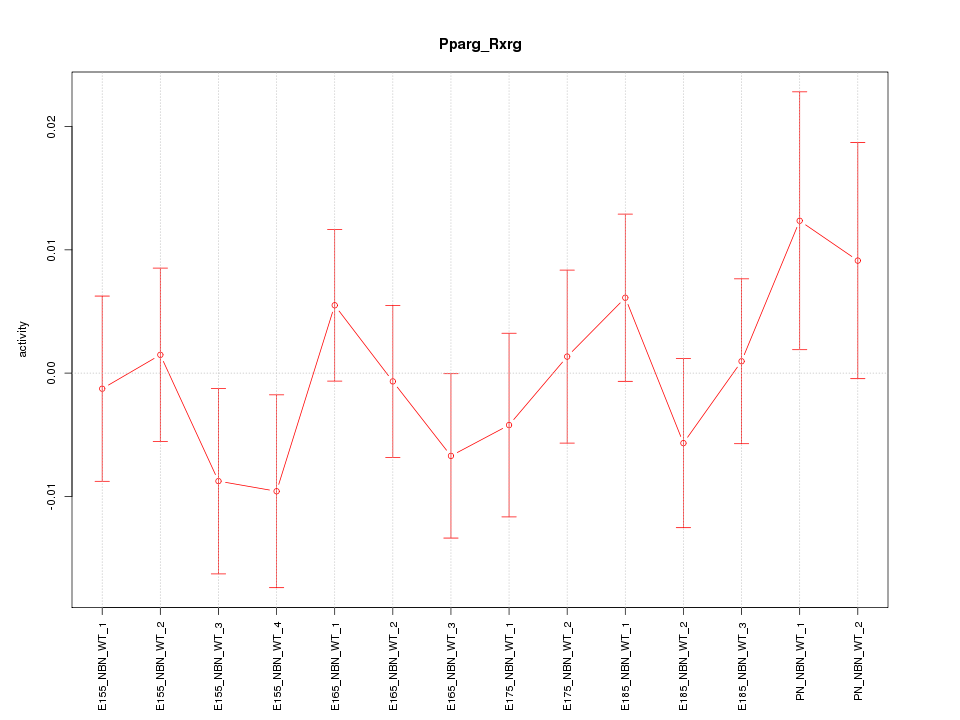
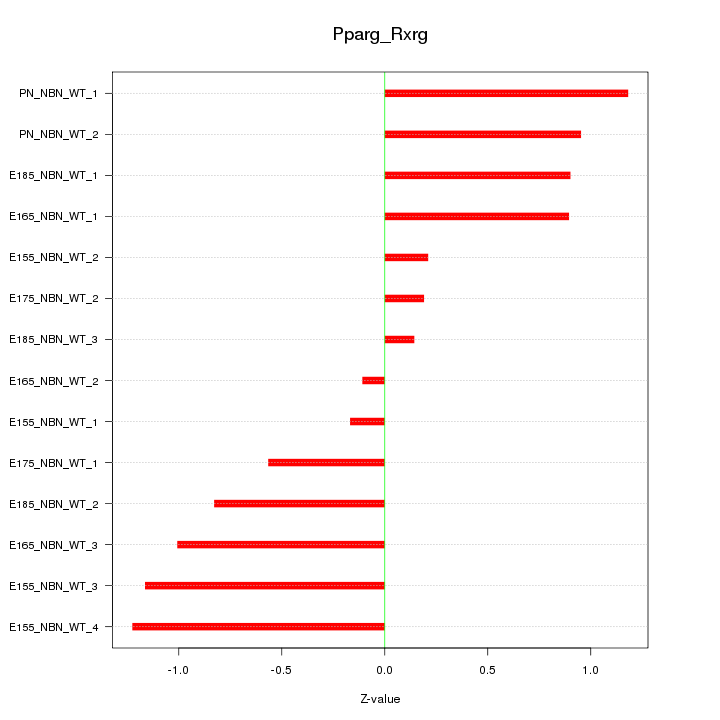
| 0.6 |
1.7 |
GO:0006553 |
lysine metabolic process(GO:0006553) |
| 0.5 |
1.4 |
GO:0014063 |
negative regulation of serotonin secretion(GO:0014063) |
| 0.4 |
1.4 |
GO:0060729 |
intestinal epithelial structure maintenance(GO:0060729) |
| 0.3 |
0.9 |
GO:0001803 |
type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 0.3 |
1.1 |
GO:0032901 |
positive regulation of neurotrophin production(GO:0032901) |
| 0.3 |
0.8 |
GO:1904685 |
positive regulation of metalloendopeptidase activity(GO:1904685) |
| 0.3 |
0.5 |
GO:0098700 |
aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.3 |
0.8 |
GO:0035582 |
sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.3 |
0.8 |
GO:2001206 |
positive regulation of osteoclast development(GO:2001206) |
| 0.2 |
0.7 |
GO:0046900 |
tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.2 |
0.6 |
GO:0006578 |
amino-acid betaine biosynthetic process(GO:0006578) |
| 0.2 |
1.1 |
GO:0010624 |
regulation of Schwann cell proliferation(GO:0010624) |
| 0.2 |
0.5 |
GO:0032918 |
polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.2 |
0.8 |
GO:0015788 |
UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.2 |
0.8 |
GO:0051365 |
cellular response to potassium ion starvation(GO:0051365) |
| 0.2 |
0.6 |
GO:1904177 |
regulation of adipose tissue development(GO:1904177) |
| 0.2 |
0.5 |
GO:1902358 |
sulfate transmembrane transport(GO:1902358) |
| 0.1 |
1.0 |
GO:0001887 |
selenium compound metabolic process(GO:0001887) |
| 0.1 |
0.4 |
GO:1903795 |
regulation of inorganic anion transmembrane transport(GO:1903795) |
| 0.1 |
0.5 |
GO:2000048 |
negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 |
0.3 |
GO:0035461 |
vitamin transmembrane transport(GO:0035461) |
| 0.1 |
0.8 |
GO:0032483 |
regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 |
1.0 |
GO:0071732 |
cellular response to nitric oxide(GO:0071732) |
| 0.1 |
0.4 |
GO:0030321 |
transepithelial chloride transport(GO:0030321) |
| 0.1 |
1.2 |
GO:0042760 |
very long-chain fatty acid catabolic process(GO:0042760) |
| 0.1 |
0.4 |
GO:0042758 |
long-chain fatty acid catabolic process(GO:0042758) |
| 0.1 |
0.4 |
GO:0043181 |
vacuolar sequestering(GO:0043181) |
| 0.1 |
0.4 |
GO:0014809 |
regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 |
0.1 |
GO:0071726 |
response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) |
| 0.1 |
0.7 |
GO:0046880 |
regulation of follicle-stimulating hormone secretion(GO:0046880) follicle-stimulating hormone secretion(GO:0046884) |
| 0.1 |
0.6 |
GO:0006659 |
phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 |
0.4 |
GO:0035701 |
hematopoietic stem cell migration(GO:0035701) |
| 0.1 |
0.6 |
GO:0000098 |
sulfur amino acid catabolic process(GO:0000098) |
| 0.1 |
0.8 |
GO:0006703 |
estrogen biosynthetic process(GO:0006703) |
| 0.1 |
0.3 |
GO:2001187 |
positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.1 |
0.2 |
GO:0071677 |
positive regulation of mononuclear cell migration(GO:0071677) |
| 0.1 |
0.3 |
GO:0008228 |
opsonization(GO:0008228) |
| 0.1 |
0.3 |
GO:1902083 |
negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 |
0.3 |
GO:0019074 |
viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 |
0.8 |
GO:0042435 |
indole-containing compound biosynthetic process(GO:0042435) |
| 0.1 |
0.4 |
GO:2001185 |
regulation of CD8-positive, alpha-beta T cell activation(GO:2001185) |
| 0.1 |
0.4 |
GO:2000427 |
positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 |
0.3 |
GO:0015920 |
lipopolysaccharide transport(GO:0015920) |
| 0.1 |
0.2 |
GO:0003096 |
renal sodium ion transport(GO:0003096) |
| 0.1 |
0.3 |
GO:0035928 |
rRNA import into mitochondrion(GO:0035928) |
| 0.1 |
0.7 |
GO:0032020 |
ISG15-protein conjugation(GO:0032020) |
| 0.1 |
0.2 |
GO:0043696 |
dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.1 |
0.4 |
GO:0010510 |
regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.1 |
0.6 |
GO:0015862 |
uridine transport(GO:0015862) |
| 0.1 |
0.3 |
GO:0048611 |
ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.1 |
0.3 |
GO:0002934 |
desmosome organization(GO:0002934) |
| 0.1 |
0.4 |
GO:1904683 |
regulation of metalloendopeptidase activity(GO:1904683) negative regulation of metalloendopeptidase activity(GO:1904684) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.1 |
0.3 |
GO:0046104 |
thymidine metabolic process(GO:0046104) |
| 0.1 |
0.3 |
GO:0002295 |
T-helper cell lineage commitment(GO:0002295) interleukin-4-mediated signaling pathway(GO:0035771) cellular response to reactive nitrogen species(GO:1902170) |
| 0.1 |
0.2 |
GO:1903998 |
regulation of eating behavior(GO:1903998) |
| 0.1 |
0.2 |
GO:0044650 |
virion attachment to host cell(GO:0019062) adhesion of symbiont to host cell(GO:0044650) |
| 0.1 |
0.6 |
GO:0071294 |
cellular response to zinc ion(GO:0071294) |
| 0.1 |
0.7 |
GO:0036158 |
outer dynein arm assembly(GO:0036158) |
| 0.1 |
1.6 |
GO:0033539 |
fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 |
0.5 |
GO:0001867 |
complement activation, lectin pathway(GO:0001867) |
| 0.1 |
0.3 |
GO:0048682 |
axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.1 |
0.2 |
GO:0070358 |
actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 |
0.2 |
GO:0031038 |
myosin II filament organization(GO:0031038) regulation of myosin II filament organization(GO:0043519) |
| 0.1 |
0.2 |
GO:0019676 |
ammonia assimilation cycle(GO:0019676) |
| 0.1 |
0.4 |
GO:0050861 |
positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 |
0.7 |
GO:0015721 |
bile acid and bile salt transport(GO:0015721) |
| 0.1 |
0.3 |
GO:0051708 |
intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) cytosol to ER transport(GO:0046967) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.1 |
0.4 |
GO:0050747 |
positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.1 |
0.5 |
GO:0051004 |
regulation of lipoprotein lipase activity(GO:0051004) |
| 0.1 |
0.3 |
GO:2000680 |
regulation of rubidium ion transport(GO:2000680) |
| 0.1 |
0.4 |
GO:0032229 |
positive regulation of gamma-aminobutyric acid secretion(GO:0014054) negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.1 |
0.3 |
GO:0051151 |
negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 |
0.2 |
GO:0035262 |
gonad morphogenesis(GO:0035262) |
| 0.1 |
0.2 |
GO:0099624 |
regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) atrial cardiac muscle cell membrane repolarization(GO:0099624) |
| 0.1 |
0.2 |
GO:2001045 |
closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 |
0.3 |
GO:0006361 |
transcription initiation from RNA polymerase I promoter(GO:0006361) termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 |
0.4 |
GO:0016557 |
peroxisome membrane biogenesis(GO:0016557) |
| 0.1 |
0.2 |
GO:0002636 |
positive regulation of germinal center formation(GO:0002636) |
| 0.1 |
0.4 |
GO:0033314 |
mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 |
0.2 |
GO:0051045 |
negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 |
0.4 |
GO:0033227 |
dsRNA transport(GO:0033227) |
| 0.1 |
0.3 |
GO:0045347 |
negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.1 |
0.3 |
GO:0071500 |
cellular response to nitrosative stress(GO:0071500) |
| 0.1 |
0.8 |
GO:1904294 |
positive regulation of ERAD pathway(GO:1904294) |
| 0.1 |
0.4 |
GO:0070314 |
G1 to G0 transition(GO:0070314) |
| 0.0 |
0.2 |
GO:0046598 |
positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 |
0.8 |
GO:0017144 |
drug metabolic process(GO:0017144) |
| 0.0 |
0.2 |
GO:0051315 |
attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 |
0.6 |
GO:0021891 |
olfactory bulb interneuron development(GO:0021891) |
| 0.0 |
0.2 |
GO:0035752 |
lysosomal lumen pH elevation(GO:0035752) |
| 0.0 |
0.8 |
GO:0006744 |
ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 |
0.1 |
GO:0086069 |
bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 |
0.6 |
GO:0000042 |
protein targeting to Golgi(GO:0000042) |
| 0.0 |
0.1 |
GO:0046655 |
folic acid metabolic process(GO:0046655) |
| 0.0 |
0.1 |
GO:0048296 |
isotype switching to IgA isotypes(GO:0048290) regulation of isotype switching to IgA isotypes(GO:0048296) |
| 0.0 |
0.5 |
GO:0016322 |
neuron remodeling(GO:0016322) |
| 0.0 |
0.2 |
GO:1903251 |
multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.0 |
0.7 |
GO:0060259 |
regulation of feeding behavior(GO:0060259) |
| 0.0 |
0.3 |
GO:1901837 |
negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.0 |
0.3 |
GO:0032929 |
negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 |
0.2 |
GO:0006438 |
valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 |
0.3 |
GO:0036462 |
TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 |
0.1 |
GO:0031999 |
negative regulation of fatty acid beta-oxidation(GO:0031999) |
| 0.0 |
0.2 |
GO:0090370 |
negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 |
0.1 |
GO:0002371 |
dendritic cell cytokine production(GO:0002371) regulation of dendritic cell cytokine production(GO:0002730) |
| 0.0 |
0.3 |
GO:0031666 |
positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 |
0.1 |
GO:0042590 |
antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.0 |
0.1 |
GO:0034310 |
primary alcohol catabolic process(GO:0034310) |
| 0.0 |
0.2 |
GO:0090272 |
negative regulation of complement activation(GO:0045916) negative regulation of fibroblast growth factor production(GO:0090272) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 |
0.2 |
GO:0019372 |
lipoxygenase pathway(GO:0019372) |
| 0.0 |
0.5 |
GO:0097034 |
mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 |
0.0 |
GO:0061349 |
planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) |
| 0.0 |
0.2 |
GO:2000491 |
positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 |
0.1 |
GO:0089700 |
protein kinase D signaling(GO:0089700) |
| 0.0 |
0.4 |
GO:0046415 |
urate metabolic process(GO:0046415) |
| 0.0 |
0.5 |
GO:0046643 |
regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.0 |
0.1 |
GO:1904192 |
cholangiocyte apoptotic process(GO:1902488) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 0.0 |
0.4 |
GO:0061032 |
visceral serous pericardium development(GO:0061032) |
| 0.0 |
0.1 |
GO:0033364 |
mast cell secretory granule organization(GO:0033364) |
| 0.0 |
0.2 |
GO:2000157 |
regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.0 |
0.1 |
GO:0061043 |
regulation of vascular wound healing(GO:0061043) |
| 0.0 |
0.4 |
GO:0048733 |
sebaceous gland development(GO:0048733) |
| 0.0 |
0.2 |
GO:0042573 |
retinoic acid metabolic process(GO:0042573) |
| 0.0 |
0.0 |
GO:0061153 |
trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 |
0.2 |
GO:0060978 |
angiogenesis involved in coronary vascular morphogenesis(GO:0060978) |
| 0.0 |
0.3 |
GO:0090050 |
positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 |
0.1 |
GO:0002484 |
antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.0 |
0.4 |
GO:0042572 |
retinol metabolic process(GO:0042572) |
| 0.0 |
0.1 |
GO:0000715 |
nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 |
0.2 |
GO:1903772 |
regulation of viral budding via host ESCRT complex(GO:1903772) |
| 0.0 |
0.7 |
GO:0008299 |
isoprenoid biosynthetic process(GO:0008299) |
| 0.0 |
0.3 |
GO:2001214 |
positive regulation of vasculogenesis(GO:2001214) |
| 0.0 |
0.1 |
GO:2000346 |
negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 |
0.2 |
GO:0033299 |
secretion of lysosomal enzymes(GO:0033299) eye pigmentation(GO:0048069) |
| 0.0 |
0.2 |
GO:0015840 |
urea transport(GO:0015840) urea transmembrane transport(GO:0071918) |
| 0.0 |
0.1 |
GO:0006421 |
asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 |
0.3 |
GO:0071493 |
cellular response to UV-B(GO:0071493) |
| 0.0 |
0.5 |
GO:2001275 |
positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 |
1.1 |
GO:0015909 |
long-chain fatty acid transport(GO:0015909) |
| 0.0 |
0.1 |
GO:0030997 |
regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 |
0.2 |
GO:0060452 |
positive regulation of cardiac muscle contraction(GO:0060452) |
| 0.0 |
0.2 |
GO:0021913 |
regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 |
0.5 |
GO:0090005 |
negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.0 |
0.7 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 |
0.7 |
GO:0050873 |
brown fat cell differentiation(GO:0050873) |
| 0.0 |
0.3 |
GO:0031274 |
positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 |
0.3 |
GO:0051988 |
regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 |
0.1 |
GO:0006287 |
base-excision repair, gap-filling(GO:0006287) |
| 0.0 |
0.0 |
GO:0086068 |
Purkinje myocyte to ventricular cardiac muscle cell signaling(GO:0086029) Purkinje myocyte to ventricular cardiac muscle cell communication(GO:0086068) |
| 0.0 |
0.2 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.0 |
0.2 |
GO:0043011 |
myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 |
0.2 |
GO:0010764 |
negative regulation of fibroblast migration(GO:0010764) |
| 0.0 |
0.2 |
GO:1902969 |
DNA replication, Okazaki fragment processing(GO:0033567) mitotic DNA replication(GO:1902969) |
| 0.0 |
0.3 |
GO:0034501 |
protein localization to kinetochore(GO:0034501) |
| 0.0 |
0.4 |
GO:0010866 |
regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.0 |
0.1 |
GO:2000544 |
cell chemotaxis to fibroblast growth factor(GO:0035766) endothelial cell chemotaxis to fibroblast growth factor(GO:0035768) regulation of cell chemotaxis to fibroblast growth factor(GO:1904847) regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000544) |
| 0.0 |
0.3 |
GO:0071803 |
positive regulation of podosome assembly(GO:0071803) |
| 0.0 |
0.1 |
GO:0042256 |
mature ribosome assembly(GO:0042256) |
| 0.0 |
0.1 |
GO:0001193 |
maintenance of transcriptional fidelity during DNA-templated transcription elongation(GO:0001192) maintenance of transcriptional fidelity during DNA-templated transcription elongation from RNA polymerase II promoter(GO:0001193) |
| 0.0 |
0.3 |
GO:0030150 |
protein import into mitochondrial matrix(GO:0030150) |
| 0.0 |
0.3 |
GO:0071257 |
cellular response to electrical stimulus(GO:0071257) |
| 0.0 |
0.1 |
GO:0021698 |
cerebellar cortex structural organization(GO:0021698) |
| 0.0 |
0.1 |
GO:0048312 |
intracellular distribution of mitochondria(GO:0048312) |
| 0.0 |
0.2 |
GO:0019368 |
fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 |
0.1 |
GO:0006669 |
sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 |
0.1 |
GO:0006478 |
peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 |
0.2 |
GO:0035845 |
photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 |
0.2 |
GO:0018344 |
protein geranylgeranylation(GO:0018344) |
| 0.0 |
0.0 |
GO:0036337 |
Fas signaling pathway(GO:0036337) |
| 0.0 |
0.3 |
GO:0014067 |
negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 |
0.1 |
GO:0035609 |
C-terminal protein deglutamylation(GO:0035609) |
| 0.0 |
0.1 |
GO:0030242 |
pexophagy(GO:0030242) |
| 0.0 |
0.3 |
GO:0000038 |
very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 |
0.1 |
GO:0042045 |
epithelial fluid transport(GO:0042045) |
| 0.0 |
0.0 |
GO:1905005 |
regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905005) |
| 0.0 |
0.1 |
GO:0009298 |
GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 |
0.5 |
GO:0045662 |
negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 |
0.8 |
GO:0071300 |
cellular response to retinoic acid(GO:0071300) |
| 0.0 |
0.3 |
GO:0032060 |
bleb assembly(GO:0032060) |
| 0.0 |
0.1 |
GO:0090611 |
ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.0 |
0.1 |
GO:0000185 |
activation of MAPKKK activity(GO:0000185) |
| 0.0 |
0.1 |
GO:0034551 |
respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 |
0.0 |
GO:0006207 |
'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 |
0.1 |
GO:0019509 |
L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 |
0.0 |
GO:0002940 |
tRNA N2-guanine methylation(GO:0002940) |
| 0.0 |
0.0 |
GO:0000430 |
regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 |
0.3 |
GO:0045947 |
negative regulation of translational initiation(GO:0045947) |
| 0.0 |
0.2 |
GO:0006268 |
DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 |
0.0 |
GO:0030497 |
fatty acid elongation(GO:0030497) |
| 0.0 |
0.1 |
GO:0032074 |
negative regulation of nuclease activity(GO:0032074) |
| 0.0 |
0.8 |
GO:0008156 |
negative regulation of DNA replication(GO:0008156) |
| 0.0 |
0.2 |
GO:0000291 |
nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.0 |
0.3 |
GO:0071392 |
cellular response to estradiol stimulus(GO:0071392) |
| 0.0 |
0.4 |
GO:0018345 |
protein palmitoylation(GO:0018345) |
| 0.0 |
0.1 |
GO:0032620 |
interleukin-17 production(GO:0032620) |
| 0.0 |
0.3 |
GO:0006491 |
N-glycan processing(GO:0006491) |
| 0.0 |
0.2 |
GO:0050901 |
leukocyte tethering or rolling(GO:0050901) |
| 0.0 |
0.1 |
GO:1903504 |
regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 |
0.0 |
GO:0046381 |
CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.0 |
0.4 |
GO:0034314 |
Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 |
0.2 |
GO:0045907 |
positive regulation of vasoconstriction(GO:0045907) |
| 0.0 |
0.1 |
GO:0016559 |
peroxisome fission(GO:0016559) |
| 0.0 |
0.3 |
GO:0000132 |
establishment of mitotic spindle orientation(GO:0000132) |
| 0.0 |
0.1 |
GO:0050765 |
negative regulation of phagocytosis(GO:0050765) |
| 0.0 |
0.1 |
GO:0043983 |
histone H4-K12 acetylation(GO:0043983) |
| 0.0 |
0.1 |
GO:0048227 |
plasma membrane to endosome transport(GO:0048227) |
| 0.0 |
0.0 |
GO:1900060 |
negative regulation of sphingolipid biosynthetic process(GO:0090155) cellular sphingolipid homeostasis(GO:0090156) negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.0 |
0.4 |
GO:0000266 |
mitochondrial fission(GO:0000266) |
| 0.0 |
0.1 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 |
0.2 |
GO:1901798 |
positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 |
0.1 |
GO:0010528 |
regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 |
0.2 |
GO:0006656 |
phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 |
0.3 |
GO:0060441 |
epithelial tube branching involved in lung morphogenesis(GO:0060441) |
| 0.0 |
0.1 |
GO:0035022 |
positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.0 |
0.1 |
GO:0007271 |
synaptic transmission, cholinergic(GO:0007271) |
| 0.0 |
0.3 |
GO:0097006 |
regulation of plasma lipoprotein particle levels(GO:0097006) |
| 0.0 |
0.1 |
GO:0045162 |
clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 |
0.4 |
GO:0045739 |
positive regulation of DNA repair(GO:0045739) |
| 0.0 |
0.1 |
GO:0007253 |
cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 |
0.0 |
GO:0070327 |
thyroid hormone transport(GO:0070327) |
| 0.0 |
0.1 |
GO:0006102 |
isocitrate metabolic process(GO:0006102) |
| 0.0 |
0.0 |
GO:0031581 |
hemidesmosome assembly(GO:0031581) |
| 0.0 |
0.1 |
GO:0071850 |
mitotic cell cycle arrest(GO:0071850) |
| 0.0 |
0.1 |
GO:0007195 |
adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 |
0.1 |
GO:0010225 |
response to UV-C(GO:0010225) |
| 0.0 |
0.1 |
GO:0030220 |
platelet formation(GO:0030220) |