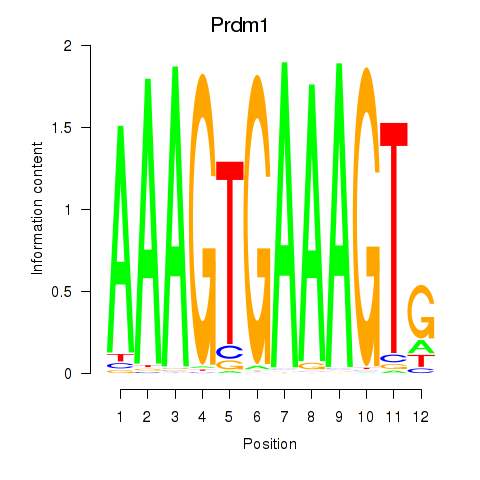
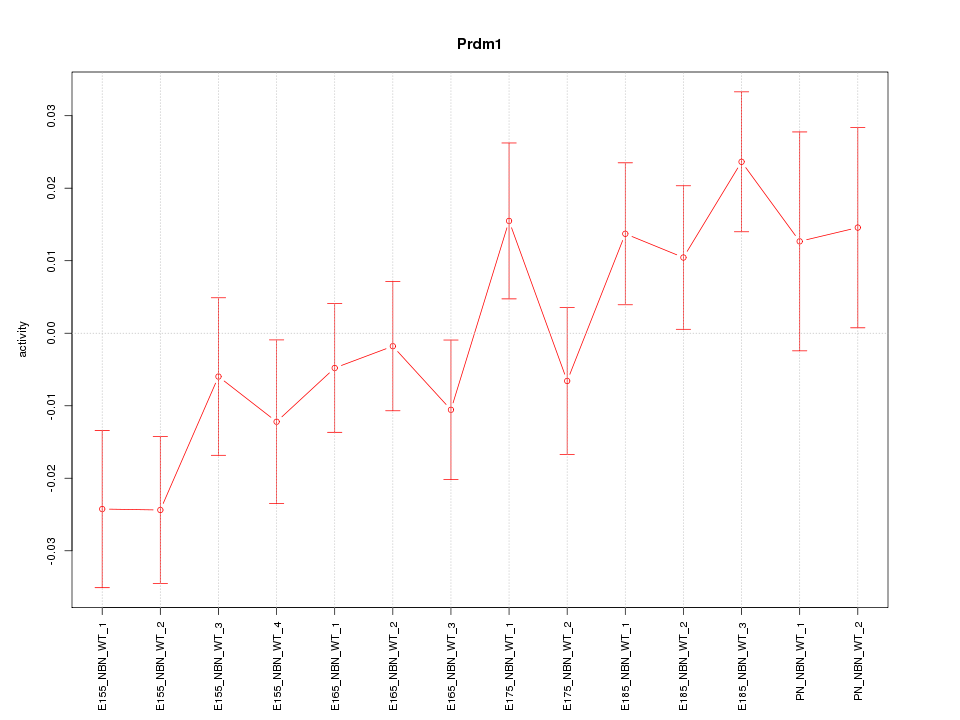
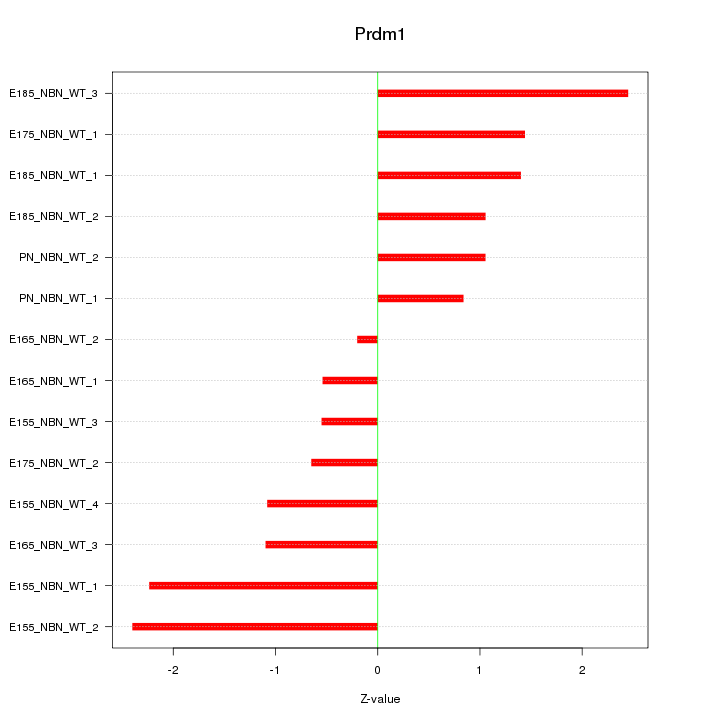
| 1.9 |
5.8 |
GO:0032765 |
positive regulation of mast cell cytokine production(GO:0032765) |
| 1.4 |
4.1 |
GO:0006001 |
fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 1.2 |
3.5 |
GO:0042196 |
dichloromethane metabolic process(GO:0018900) chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 0.7 |
2.1 |
GO:0061537 |
glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.7 |
4.7 |
GO:0090336 |
positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.6 |
3.7 |
GO:0003383 |
apical constriction(GO:0003383) |
| 0.6 |
2.5 |
GO:0035934 |
corticosterone secretion(GO:0035934) |
| 0.6 |
1.7 |
GO:0045349 |
interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.5 |
1.6 |
GO:0043181 |
vacuolar sequestering(GO:0043181) |
| 0.5 |
2.0 |
GO:1903984 |
positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.4 |
1.8 |
GO:0006867 |
asparagine transport(GO:0006867) glutamine transport(GO:0006868) |
| 0.4 |
1.7 |
GO:0070358 |
actin polymerization-dependent cell motility(GO:0070358) |
| 0.4 |
1.6 |
GO:1903416 |
response to glycoside(GO:1903416) |
| 0.4 |
1.5 |
GO:1903237 |
negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.4 |
1.1 |
GO:2001180 |
negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.3 |
0.9 |
GO:1902915 |
negative regulation of protein import into nucleus, translocation(GO:0033159) protein poly-ADP-ribosylation(GO:0070212) negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.3 |
0.9 |
GO:1904139 |
microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.2 |
1.8 |
GO:0021984 |
adenohypophysis development(GO:0021984) |
| 0.2 |
0.9 |
GO:0030951 |
establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.2 |
0.5 |
GO:1990481 |
snoRNA guided rRNA pseudouridine synthesis(GO:0000454) snRNA pseudouridine synthesis(GO:0031120) mRNA pseudouridine synthesis(GO:1990481) |
| 0.2 |
0.9 |
GO:0010701 |
positive regulation of norepinephrine secretion(GO:0010701) |
| 0.2 |
1.0 |
GO:0006957 |
complement activation, alternative pathway(GO:0006957) |
| 0.2 |
2.3 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.2 |
1.0 |
GO:0042590 |
antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.1 |
1.3 |
GO:0042760 |
very long-chain fatty acid catabolic process(GO:0042760) |
| 0.1 |
0.6 |
GO:0072592 |
regulation of integrin biosynthetic process(GO:0045113) oxygen metabolic process(GO:0072592) |
| 0.1 |
1.2 |
GO:0030432 |
peristalsis(GO:0030432) |
| 0.1 |
1.9 |
GO:2001275 |
positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.1 |
0.3 |
GO:1904879 |
positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.1 |
0.3 |
GO:0021546 |
rhombomere development(GO:0021546) |
| 0.1 |
0.3 |
GO:0010757 |
arachidonic acid metabolite production involved in inflammatory response(GO:0002538) negative regulation of plasminogen activation(GO:0010757) regulation of vascular wound healing(GO:0061043) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 |
0.6 |
GO:0051694 |
pointed-end actin filament capping(GO:0051694) |
| 0.1 |
0.3 |
GO:0045404 |
positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.1 |
3.3 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.1 |
1.3 |
GO:0021902 |
commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 |
0.2 |
GO:0014873 |
response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.1 |
0.3 |
GO:1902990 |
mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.1 |
0.2 |
GO:0046077 |
dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.1 |
1.6 |
GO:0006828 |
manganese ion transport(GO:0006828) |
| 0.1 |
1.4 |
GO:0042832 |
defense response to protozoan(GO:0042832) |
| 0.1 |
0.3 |
GO:1903802 |
positive regulation of arachidonic acid secretion(GO:0090238) L-glutamate(1-) import into cell(GO:1903802) L-glutamate import into cell(GO:1990123) |
| 0.1 |
0.3 |
GO:0010756 |
positive regulation of plasminogen activation(GO:0010756) |
| 0.1 |
0.4 |
GO:0002317 |
plasma cell differentiation(GO:0002317) |
| 0.1 |
0.6 |
GO:0006450 |
regulation of translational fidelity(GO:0006450) |
| 0.1 |
0.5 |
GO:0048625 |
myoblast fate commitment(GO:0048625) |
| 0.1 |
0.8 |
GO:0050930 |
induction of positive chemotaxis(GO:0050930) |
| 0.1 |
0.2 |
GO:0019885 |
antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 |
0.2 |
GO:2001274 |
negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.1 |
0.6 |
GO:0030322 |
stabilization of membrane potential(GO:0030322) |
| 0.1 |
0.3 |
GO:0045716 |
positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.1 |
0.9 |
GO:0071577 |
zinc II ion transmembrane transport(GO:0071577) |
| 0.1 |
0.4 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 |
0.7 |
GO:0019243 |
methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 |
0.4 |
GO:0071907 |
determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.0 |
0.3 |
GO:2000270 |
negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 |
1.3 |
GO:0046677 |
response to antibiotic(GO:0046677) |
| 0.0 |
0.7 |
GO:0043248 |
proteasome assembly(GO:0043248) |
| 0.0 |
0.1 |
GO:0006553 |
lysine metabolic process(GO:0006553) |
| 0.0 |
0.3 |
GO:0043383 |
negative T cell selection(GO:0043383) negative thymic T cell selection(GO:0045060) |
| 0.0 |
0.3 |
GO:0032020 |
ISG15-protein conjugation(GO:0032020) |
| 0.0 |
0.1 |
GO:0006407 |
rRNA export from nucleus(GO:0006407) |
| 0.0 |
0.3 |
GO:0090370 |
negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 |
0.9 |
GO:2000811 |
negative regulation of anoikis(GO:2000811) |
| 0.0 |
0.7 |
GO:1900027 |
regulation of ruffle assembly(GO:1900027) |
| 0.0 |
0.6 |
GO:0006465 |
signal peptide processing(GO:0006465) |
| 0.0 |
0.4 |
GO:0048672 |
positive regulation of collateral sprouting(GO:0048672) |
| 0.0 |
0.1 |
GO:0046598 |
positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 |
0.4 |
GO:2000696 |
regulation of epithelial cell differentiation involved in kidney development(GO:2000696) |
| 0.0 |
0.1 |
GO:0045448 |
regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.0 |
0.1 |
GO:0043696 |
dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.0 |
0.4 |
GO:0006268 |
DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 |
0.1 |
GO:1901069 |
guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 |
0.2 |
GO:0018401 |
peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 |
0.6 |
GO:0019884 |
antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.0 |
0.2 |
GO:0042126 |
nitrate metabolic process(GO:0042126) |
| 0.0 |
0.2 |
GO:0032782 |
bile acid secretion(GO:0032782) |
| 0.0 |
0.4 |
GO:0006744 |
ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 |
0.3 |
GO:0014842 |
regulation of skeletal muscle satellite cell proliferation(GO:0014842) regulation of skeletal muscle cell proliferation(GO:0014857) |
| 0.0 |
0.2 |
GO:0006451 |
selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 |
0.1 |
GO:0060050 |
positive regulation of protein glycosylation(GO:0060050) |
| 0.0 |
0.4 |
GO:2001022 |
positive regulation of response to DNA damage stimulus(GO:2001022) |
| 0.0 |
2.7 |
GO:0048814 |
regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 |
0.2 |
GO:1990403 |
embryonic brain development(GO:1990403) |
| 0.0 |
0.1 |
GO:1904721 |
regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) regulation of endoribonuclease activity(GO:0060699) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 |
0.1 |
GO:0046826 |
negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 |
1.3 |
GO:0034644 |
cellular response to UV(GO:0034644) |
| 0.0 |
0.7 |
GO:0006284 |
base-excision repair(GO:0006284) |
| 0.0 |
2.0 |
GO:0016052 |
carbohydrate catabolic process(GO:0016052) |
| 0.0 |
0.0 |
GO:0002014 |
vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.0 |
0.1 |
GO:0000491 |
small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 |
0.5 |
GO:0070098 |
chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 |
0.8 |
GO:0070527 |
platelet aggregation(GO:0070527) |
| 0.0 |
1.2 |
GO:0051896 |
regulation of protein kinase B signaling(GO:0051896) |
| 0.0 |
0.7 |
GO:0032543 |
mitochondrial translation(GO:0032543) |
| 0.0 |
0.1 |
GO:1900194 |
negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 |
0.1 |
GO:0006614 |
SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |