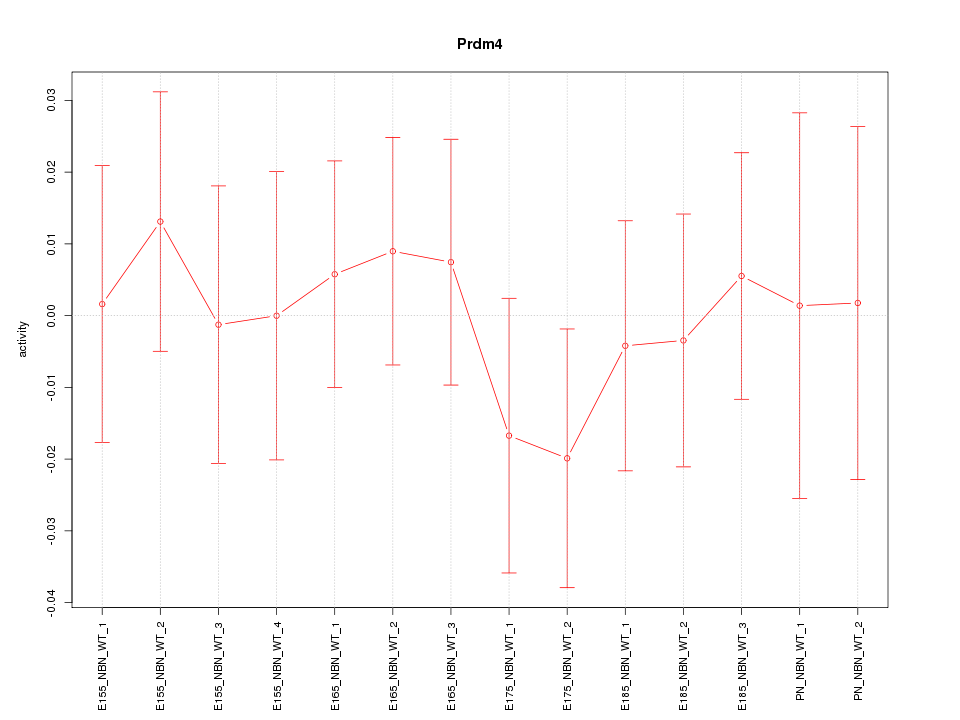
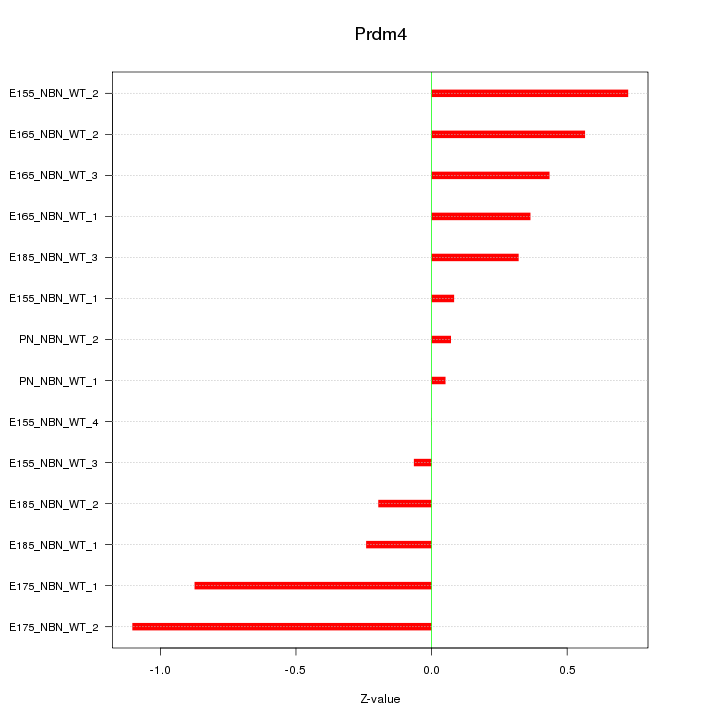
Motif ID: Prdm4
Z-value: 0.491
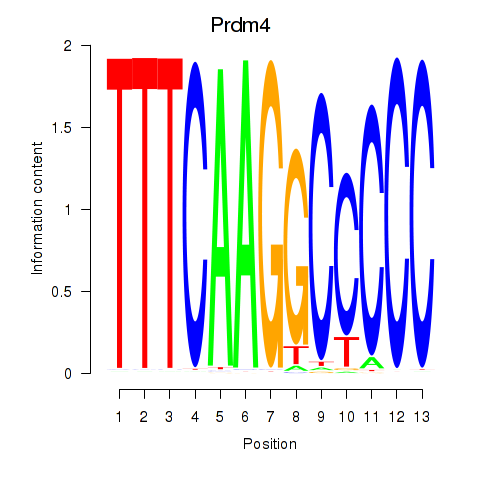
Transcription factors associated with Prdm4:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Prdm4 | ENSMUSG00000035529.9 | Prdm4 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Prdm4 | mm10_v2_chr10_-_85916902_85917006 | 0.27 | 3.6e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.0 | GO:0003150 | muscular septum morphogenesis(GO:0003150) |
| 0.0 | 0.4 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.0 | 0.5 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.2 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.0 | 0.6 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 1.4 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.4 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 2.0 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.6 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |