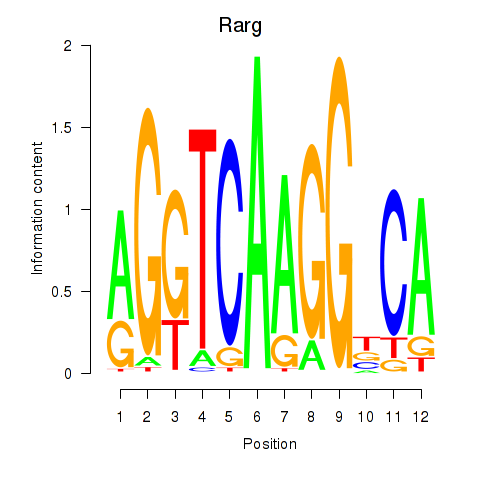
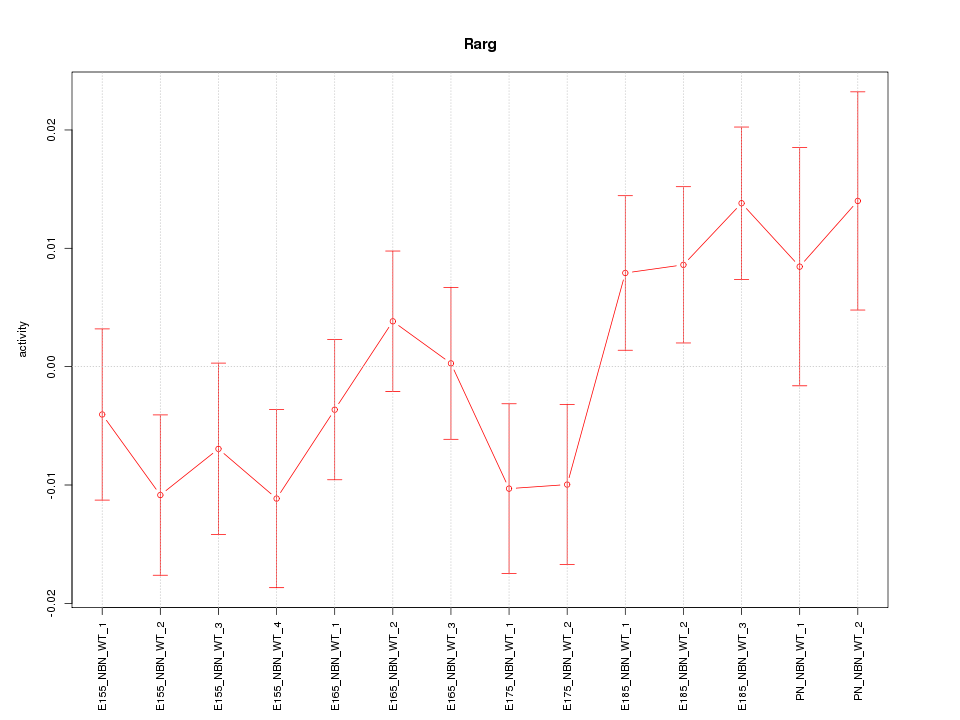
| 2.1 |
6.3 |
GO:0001803 |
antibody-dependent cellular cytotoxicity(GO:0001788) type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 0.6 |
3.1 |
GO:1904426 |
positive regulation of GTP binding(GO:1904426) |
| 0.4 |
2.6 |
GO:0036233 |
glycine import(GO:0036233) |
| 0.4 |
1.7 |
GO:0010813 |
neuropeptide catabolic process(GO:0010813) dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.4 |
1.3 |
GO:0009177 |
deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) |
| 0.4 |
1.3 |
GO:1900673 |
olefin metabolic process(GO:1900673) |
| 0.4 |
1.1 |
GO:1902358 |
sulfate transmembrane transport(GO:1902358) |
| 0.3 |
2.0 |
GO:2000672 |
negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.3 |
0.9 |
GO:0010636 |
positive regulation of mitochondrial fusion(GO:0010636) |
| 0.3 |
1.2 |
GO:0032489 |
regulation of Cdc42 protein signal transduction(GO:0032489) platelet dense granule organization(GO:0060155) |
| 0.3 |
0.8 |
GO:0006296 |
base-excision repair, AP site formation(GO:0006285) nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.2 |
1.4 |
GO:0098543 |
detection of bacterium(GO:0016045) negative regulation of natural killer cell activation(GO:0032815) detection of other organism(GO:0098543) |
| 0.2 |
1.0 |
GO:0006867 |
asparagine transport(GO:0006867) glutamine transport(GO:0006868) |
| 0.2 |
1.0 |
GO:0009414 |
response to water deprivation(GO:0009414) |
| 0.2 |
1.2 |
GO:0033136 |
serine phosphorylation of STAT3 protein(GO:0033136) |
| 0.2 |
1.6 |
GO:0015862 |
uridine transport(GO:0015862) |
| 0.2 |
1.4 |
GO:0034316 |
negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.2 |
0.7 |
GO:0010757 |
negative regulation of plasminogen activation(GO:0010757) |
| 0.2 |
0.6 |
GO:0043181 |
vacuolar sequestering(GO:0043181) |
| 0.2 |
0.4 |
GO:0002606 |
dendritic cell antigen processing and presentation(GO:0002468) regulation of dendritic cell antigen processing and presentation(GO:0002604) positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.2 |
1.5 |
GO:0019254 |
carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.2 |
0.8 |
GO:0006547 |
histidine metabolic process(GO:0006547) |
| 0.2 |
0.8 |
GO:0015888 |
thiamine transport(GO:0015888) |
| 0.2 |
1.3 |
GO:0034638 |
phosphatidylcholine catabolic process(GO:0034638) |
| 0.2 |
0.5 |
GO:1902037 |
positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.2 |
0.9 |
GO:0070305 |
response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) |
| 0.2 |
1.2 |
GO:0090336 |
positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.2 |
0.2 |
GO:2000823 |
regulation of androgen receptor activity(GO:2000823) |
| 0.2 |
1.0 |
GO:0043415 |
positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.2 |
0.8 |
GO:2000298 |
regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.2 |
0.8 |
GO:2000680 |
regulation of rubidium ion transport(GO:2000680) |
| 0.2 |
0.6 |
GO:0000379 |
tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.2 |
0.5 |
GO:0060455 |
negative regulation of gastric acid secretion(GO:0060455) |
| 0.1 |
0.4 |
GO:0060120 |
auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.1 |
0.4 |
GO:0042364 |
water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.1 |
0.4 |
GO:1901552 |
positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 |
0.4 |
GO:0046900 |
tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.1 |
0.4 |
GO:2000642 |
negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.1 |
0.4 |
GO:0010898 |
positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.1 |
0.6 |
GO:0042796 |
snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 |
0.4 |
GO:0006269 |
DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 |
0.5 |
GO:0010510 |
regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.1 |
1.1 |
GO:0038028 |
insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 |
0.4 |
GO:0007084 |
mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 |
0.4 |
GO:0003100 |
regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) |
| 0.1 |
0.3 |
GO:0090503 |
RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.1 |
0.4 |
GO:0046098 |
purine nucleobase salvage(GO:0043096) guanine metabolic process(GO:0046098) hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.1 |
0.4 |
GO:0046544 |
development of secondary male sexual characteristics(GO:0046544) |
| 0.1 |
0.4 |
GO:0042822 |
vitamin B6 metabolic process(GO:0042816) pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.1 |
1.5 |
GO:0032688 |
negative regulation of interferon-beta production(GO:0032688) |
| 0.1 |
1.2 |
GO:0043206 |
extracellular fibril organization(GO:0043206) |
| 0.1 |
0.4 |
GO:0016598 |
protein arginylation(GO:0016598) |
| 0.1 |
1.0 |
GO:0060576 |
intestinal epithelial cell development(GO:0060576) |
| 0.1 |
0.1 |
GO:0036324 |
vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.1 |
0.6 |
GO:0002457 |
T cell antigen processing and presentation(GO:0002457) |
| 0.1 |
0.9 |
GO:0018401 |
peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 |
0.3 |
GO:0032847 |
positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of cellular pH reduction(GO:0032847) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 |
0.3 |
GO:0006545 |
glycine biosynthetic process(GO:0006545) |
| 0.1 |
0.4 |
GO:0060830 |
ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.1 |
1.3 |
GO:0016322 |
neuron remodeling(GO:0016322) |
| 0.1 |
1.6 |
GO:0042759 |
long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 |
0.3 |
GO:0050713 |
negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.1 |
0.8 |
GO:0002862 |
negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.1 |
0.7 |
GO:0009449 |
gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.1 |
1.2 |
GO:0033623 |
regulation of integrin activation(GO:0033623) |
| 0.1 |
0.3 |
GO:0097461 |
ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) |
| 0.1 |
0.9 |
GO:0010820 |
positive regulation of T cell chemotaxis(GO:0010820) |
| 0.1 |
0.4 |
GO:0070589 |
cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) cell wall biogenesis(GO:0042546) cell wall macromolecule metabolic process(GO:0044036) cell wall macromolecule biosynthetic process(GO:0044038) chain elongation of O-linked mannose residue(GO:0044845) cellular component macromolecule biosynthetic process(GO:0070589) cell wall organization or biogenesis(GO:0071554) |
| 0.1 |
0.3 |
GO:0042732 |
D-xylose metabolic process(GO:0042732) |
| 0.1 |
0.6 |
GO:0006957 |
complement activation, alternative pathway(GO:0006957) |
| 0.1 |
0.1 |
GO:1902460 |
regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.1 |
0.4 |
GO:0033088 |
negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.1 |
0.5 |
GO:1903772 |
regulation of viral budding via host ESCRT complex(GO:1903772) |
| 0.1 |
0.9 |
GO:1901409 |
regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 |
0.5 |
GO:0042167 |
heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 |
0.3 |
GO:0019478 |
D-amino acid catabolic process(GO:0019478) |
| 0.1 |
0.8 |
GO:0045602 |
negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.1 |
0.3 |
GO:0014877 |
response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 |
0.5 |
GO:0001980 |
regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.1 |
0.2 |
GO:0007181 |
transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.1 |
0.6 |
GO:0097460 |
ferrous iron import into cell(GO:0097460) |
| 0.1 |
0.4 |
GO:2001268 |
negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.1 |
0.6 |
GO:0045059 |
positive thymic T cell selection(GO:0045059) |
| 0.1 |
2.3 |
GO:0071353 |
cellular response to interleukin-4(GO:0071353) |
| 0.1 |
0.4 |
GO:1900186 |
negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.1 |
0.2 |
GO:0006566 |
threonine metabolic process(GO:0006566) |
| 0.1 |
0.3 |
GO:0032417 |
positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.1 |
0.2 |
GO:1901228 |
positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.1 |
1.0 |
GO:0019243 |
methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 |
0.4 |
GO:0032929 |
negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 |
0.8 |
GO:0048733 |
sebaceous gland development(GO:0048733) |
| 0.1 |
0.2 |
GO:0042535 |
positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.1 |
0.4 |
GO:1903215 |
negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.1 |
0.7 |
GO:0039702 |
viral budding via host ESCRT complex(GO:0039702) |
| 0.1 |
0.2 |
GO:0051031 |
tRNA transport(GO:0051031) |
| 0.1 |
0.2 |
GO:0001866 |
NK T cell proliferation(GO:0001866) |
| 0.1 |
1.0 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 |
1.3 |
GO:0016558 |
protein import into peroxisome matrix(GO:0016558) |
| 0.1 |
0.4 |
GO:0044829 |
positive regulation by host of viral genome replication(GO:0044829) |
| 0.1 |
0.2 |
GO:0038203 |
TORC2 signaling(GO:0038203) |
| 0.1 |
0.5 |
GO:1902166 |
negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.1 |
0.2 |
GO:1901420 |
negative regulation of response to alcohol(GO:1901420) |
| 0.1 |
0.2 |
GO:0014874 |
response to muscle activity involved in regulation of muscle adaptation(GO:0014873) response to stimulus involved in regulation of muscle adaptation(GO:0014874) |
| 0.1 |
0.7 |
GO:0045607 |
regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 |
0.2 |
GO:0006558 |
L-phenylalanine metabolic process(GO:0006558) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) |
| 0.1 |
0.3 |
GO:2000383 |
regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.1 |
1.6 |
GO:1904355 |
positive regulation of telomere capping(GO:1904355) |
| 0.1 |
0.5 |
GO:0051534 |
negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 |
0.7 |
GO:0007100 |
mitotic centrosome separation(GO:0007100) |
| 0.1 |
0.2 |
GO:1903054 |
negative regulation of extracellular matrix organization(GO:1903054) |
| 0.1 |
0.6 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.1 |
0.6 |
GO:1902254 |
negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.1 |
1.0 |
GO:0032757 |
positive regulation of interleukin-8 production(GO:0032757) |
| 0.1 |
1.4 |
GO:0031065 |
positive regulation of histone deacetylation(GO:0031065) |
| 0.1 |
0.3 |
GO:0006348 |
chromatin silencing at telomere(GO:0006348) |
| 0.1 |
0.2 |
GO:1902512 |
positive regulation of apoptotic DNA fragmentation(GO:1902512) positive regulation of DNA catabolic process(GO:1903626) |
| 0.1 |
1.1 |
GO:0043567 |
regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 |
0.6 |
GO:1902231 |
positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.1 |
0.3 |
GO:0038018 |
Wnt receptor catabolic process(GO:0038018) |
| 0.1 |
0.6 |
GO:0045647 |
negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 |
0.4 |
GO:0006451 |
selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 |
0.8 |
GO:0043402 |
glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 |
0.2 |
GO:0035106 |
operant conditioning(GO:0035106) |
| 0.1 |
0.2 |
GO:0090370 |
negative regulation of cholesterol efflux(GO:0090370) |
| 0.1 |
0.3 |
GO:0060385 |
axonogenesis involved in innervation(GO:0060385) |
| 0.1 |
0.3 |
GO:0045605 |
negative regulation of epidermal cell differentiation(GO:0045605) |
| 0.1 |
0.3 |
GO:0070816 |
phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 |
0.3 |
GO:2000324 |
positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 |
0.1 |
GO:1902837 |
amino acid import into cell(GO:1902837) |
| 0.1 |
0.2 |
GO:0000715 |
nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.1 |
0.4 |
GO:0045199 |
maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 |
0.2 |
GO:0034983 |
peptidyl-lysine deacetylation(GO:0034983) |
| 0.1 |
0.3 |
GO:0051973 |
positive regulation of telomerase activity(GO:0051973) |
| 0.0 |
0.4 |
GO:1900262 |
regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 |
0.3 |
GO:0090037 |
positive regulation of protein kinase C signaling(GO:0090037) |
| 0.0 |
0.3 |
GO:0045200 |
establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 |
0.2 |
GO:1902626 |
assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 |
0.6 |
GO:0010388 |
cullin deneddylation(GO:0010388) |
| 0.0 |
0.3 |
GO:0035999 |
tetrahydrofolate interconversion(GO:0035999) |
| 0.0 |
0.2 |
GO:0045039 |
protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 |
0.9 |
GO:0001504 |
neurotransmitter uptake(GO:0001504) |
| 0.0 |
0.2 |
GO:0016127 |
cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 |
0.6 |
GO:0002828 |
regulation of type 2 immune response(GO:0002828) |
| 0.0 |
0.4 |
GO:0003215 |
cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 |
0.3 |
GO:0060040 |
retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 |
0.4 |
GO:0036158 |
outer dynein arm assembly(GO:0036158) |
| 0.0 |
1.2 |
GO:0008299 |
isoprenoid biosynthetic process(GO:0008299) |
| 0.0 |
0.2 |
GO:0032364 |
oxygen homeostasis(GO:0032364) |
| 0.0 |
0.3 |
GO:0045198 |
establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 |
0.2 |
GO:1901894 |
regulation of calcium-transporting ATPase activity(GO:1901894) |
| 0.0 |
0.3 |
GO:0070234 |
positive regulation of T cell apoptotic process(GO:0070234) |
| 0.0 |
0.3 |
GO:0048227 |
plasma membrane to endosome transport(GO:0048227) |
| 0.0 |
0.7 |
GO:0048240 |
sperm capacitation(GO:0048240) |
| 0.0 |
0.3 |
GO:0035881 |
amacrine cell differentiation(GO:0035881) |
| 0.0 |
0.1 |
GO:1902445 |
regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.0 |
0.2 |
GO:0007343 |
egg activation(GO:0007343) |
| 0.0 |
0.6 |
GO:0034122 |
negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.0 |
0.2 |
GO:0045900 |
negative regulation of translational elongation(GO:0045900) |
| 0.0 |
0.3 |
GO:0046085 |
adenosine metabolic process(GO:0046085) |
| 0.0 |
0.5 |
GO:0030150 |
protein import into mitochondrial matrix(GO:0030150) |
| 0.0 |
0.6 |
GO:0045943 |
positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 |
0.2 |
GO:0051661 |
maintenance of centrosome location(GO:0051661) |
| 0.0 |
0.5 |
GO:0034389 |
lipid particle organization(GO:0034389) |
| 0.0 |
0.1 |
GO:0017183 |
peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 |
0.2 |
GO:0090666 |
scaRNA localization to Cajal body(GO:0090666) |
| 0.0 |
0.4 |
GO:1904814 |
regulation of protein localization to chromosome, telomeric region(GO:1904814) |
| 0.0 |
0.1 |
GO:0070682 |
proteasome regulatory particle assembly(GO:0070682) |
| 0.0 |
0.1 |
GO:0006438 |
valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 |
0.6 |
GO:0006515 |
misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 |
0.4 |
GO:0001731 |
formation of translation preinitiation complex(GO:0001731) |
| 0.0 |
0.2 |
GO:0071806 |
intracellular protein transmembrane transport(GO:0065002) protein transmembrane transport(GO:0071806) |
| 0.0 |
0.1 |
GO:0070213 |
protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 |
0.6 |
GO:0045653 |
negative regulation of megakaryocyte differentiation(GO:0045653) negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.0 |
0.1 |
GO:0046032 |
ADP catabolic process(GO:0046032) |
| 0.0 |
0.1 |
GO:0034214 |
protein hexamerization(GO:0034214) |
| 0.0 |
0.3 |
GO:0050965 |
detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 |
0.2 |
GO:0006977 |
DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 |
0.1 |
GO:0060744 |
thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 |
0.3 |
GO:2000251 |
positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 |
0.2 |
GO:0006102 |
isocitrate metabolic process(GO:0006102) |
| 0.0 |
0.1 |
GO:0034287 |
detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 |
0.3 |
GO:0045063 |
T-helper 1 cell differentiation(GO:0045063) |
| 0.0 |
1.2 |
GO:0045600 |
positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 |
0.2 |
GO:0035330 |
regulation of hippo signaling(GO:0035330) |
| 0.0 |
0.3 |
GO:0006122 |
mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 |
0.3 |
GO:0019262 |
N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 |
0.2 |
GO:0000103 |
sulfate assimilation(GO:0000103) |
| 0.0 |
0.2 |
GO:0006646 |
phosphatidylethanolamine biosynthetic process(GO:0006646) phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 |
0.1 |
GO:1903862 |
positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 |
0.1 |
GO:0070327 |
thyroid hormone transport(GO:0070327) |
| 0.0 |
0.1 |
GO:0008612 |
peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 |
0.1 |
GO:1990034 |
calcium ion export from cell(GO:1990034) |
| 0.0 |
0.3 |
GO:0006090 |
pyruvate metabolic process(GO:0006090) |
| 0.0 |
0.4 |
GO:0043249 |
erythrocyte maturation(GO:0043249) |
| 0.0 |
1.0 |
GO:0051496 |
positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 |
0.6 |
GO:0007271 |
synaptic transmission, cholinergic(GO:0007271) |
| 0.0 |
0.6 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 |
0.4 |
GO:0051044 |
positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 |
0.0 |
GO:0010725 |
regulation of primitive erythrocyte differentiation(GO:0010725) |
| 0.0 |
0.4 |
GO:0006465 |
signal peptide processing(GO:0006465) |
| 0.0 |
0.2 |
GO:0002082 |
regulation of oxidative phosphorylation(GO:0002082) |
| 0.0 |
0.6 |
GO:0006270 |
DNA replication initiation(GO:0006270) |
| 0.0 |
0.2 |
GO:0006120 |
mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 |
0.2 |
GO:0006020 |
inositol metabolic process(GO:0006020) |
| 0.0 |
0.9 |
GO:0032784 |
regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 |
0.1 |
GO:0014886 |
transition between slow and fast fiber(GO:0014886) |
| 0.0 |
0.1 |
GO:1904948 |
midbrain dopaminergic neuron differentiation(GO:1904948) |
| 0.0 |
0.5 |
GO:0030513 |
positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 |
0.3 |
GO:0032060 |
bleb assembly(GO:0032060) |
| 0.0 |
0.2 |
GO:0009072 |
aromatic amino acid family metabolic process(GO:0009072) |
| 0.0 |
0.1 |
GO:0010587 |
miRNA catabolic process(GO:0010587) |
| 0.0 |
0.2 |
GO:0050432 |
catecholamine secretion(GO:0050432) regulation of catecholamine secretion(GO:0050433) |
| 0.0 |
0.2 |
GO:1904587 |
glycoprotein ERAD pathway(GO:0097466) response to glycoprotein(GO:1904587) |
| 0.0 |
0.3 |
GO:0043457 |
regulation of cellular respiration(GO:0043457) |
| 0.0 |
0.2 |
GO:0015868 |
purine nucleotide transport(GO:0015865) purine ribonucleotide transport(GO:0015868) |
| 0.0 |
0.1 |
GO:0015910 |
peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 |
0.7 |
GO:0006418 |
tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 |
0.1 |
GO:0009249 |
protein lipoylation(GO:0009249) |
| 0.0 |
0.1 |
GO:0031145 |
anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.0 |
0.6 |
GO:0005978 |
glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 |
0.1 |
GO:0016340 |
calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.0 |
0.8 |
GO:0008156 |
negative regulation of DNA replication(GO:0008156) |
| 0.0 |
0.4 |
GO:0008089 |
anterograde axonal transport(GO:0008089) |
| 0.0 |
0.3 |
GO:0040034 |
regulation of development, heterochronic(GO:0040034) |
| 0.0 |
0.1 |
GO:0034454 |
microtubule anchoring at centrosome(GO:0034454) |
| 0.0 |
0.0 |
GO:0043456 |
regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 |
0.1 |
GO:0061436 |
establishment of skin barrier(GO:0061436) |
| 0.0 |
0.6 |
GO:0007566 |
embryo implantation(GO:0007566) |
| 0.0 |
0.1 |
GO:0006390 |
transcription from mitochondrial promoter(GO:0006390) |
| 0.0 |
0.2 |
GO:0032515 |
negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 |
0.1 |
GO:0000185 |
activation of MAPKKK activity(GO:0000185) |
| 0.0 |
0.3 |
GO:0097320 |
membrane tubulation(GO:0097320) |
| 0.0 |
0.3 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 |
0.1 |
GO:0034227 |
tRNA thio-modification(GO:0034227) |
| 0.0 |
0.2 |
GO:0046325 |
negative regulation of glucose import(GO:0046325) |
| 0.0 |
0.4 |
GO:0006829 |
zinc II ion transport(GO:0006829) |
| 0.0 |
0.2 |
GO:0030213 |
hyaluronan biosynthetic process(GO:0030213) |
| 0.0 |
0.1 |
GO:1903887 |
motile primary cilium assembly(GO:1903887) protein localization to ciliary transition zone(GO:1904491) |
| 0.0 |
0.1 |
GO:0030206 |
chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 |
0.0 |
GO:0014047 |
glutamate secretion(GO:0014047) |
| 0.0 |
0.4 |
GO:0000266 |
mitochondrial fission(GO:0000266) |
| 0.0 |
0.3 |
GO:0001829 |
trophectodermal cell differentiation(GO:0001829) |
| 0.0 |
0.3 |
GO:0048025 |
negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 |
0.2 |
GO:0043551 |
regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 |
0.1 |
GO:0006968 |
cellular defense response(GO:0006968) |
| 0.0 |
0.4 |
GO:0045843 |
negative regulation of striated muscle tissue development(GO:0045843) negative regulation of muscle tissue development(GO:1901862) |
| 0.0 |
0.3 |
GO:0046677 |
response to antibiotic(GO:0046677) |
| 0.0 |
0.1 |
GO:2000059 |
negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.0 |
0.0 |
GO:0070345 |
negative regulation of fat cell proliferation(GO:0070345) |