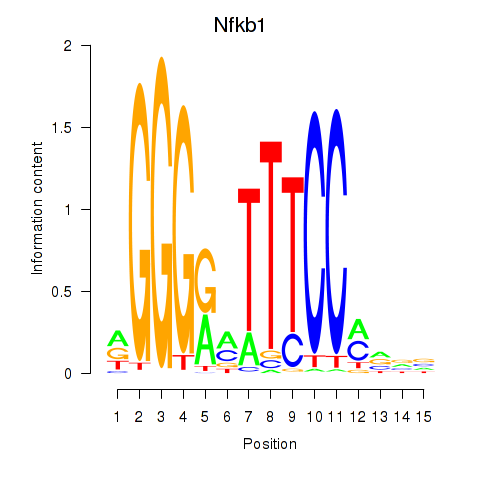
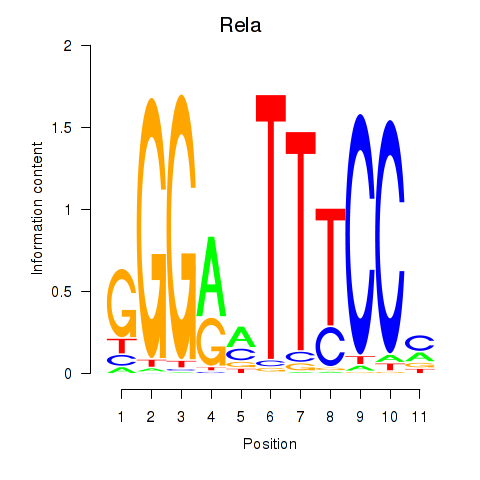
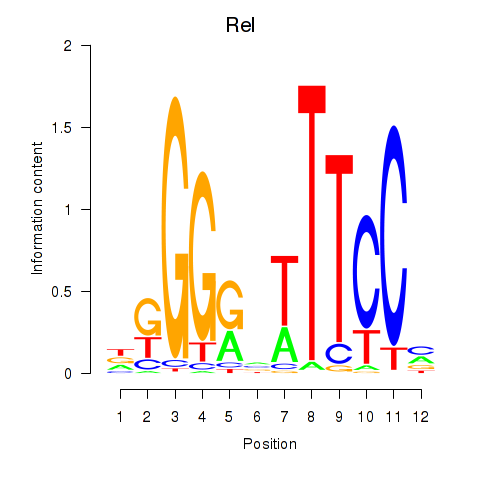
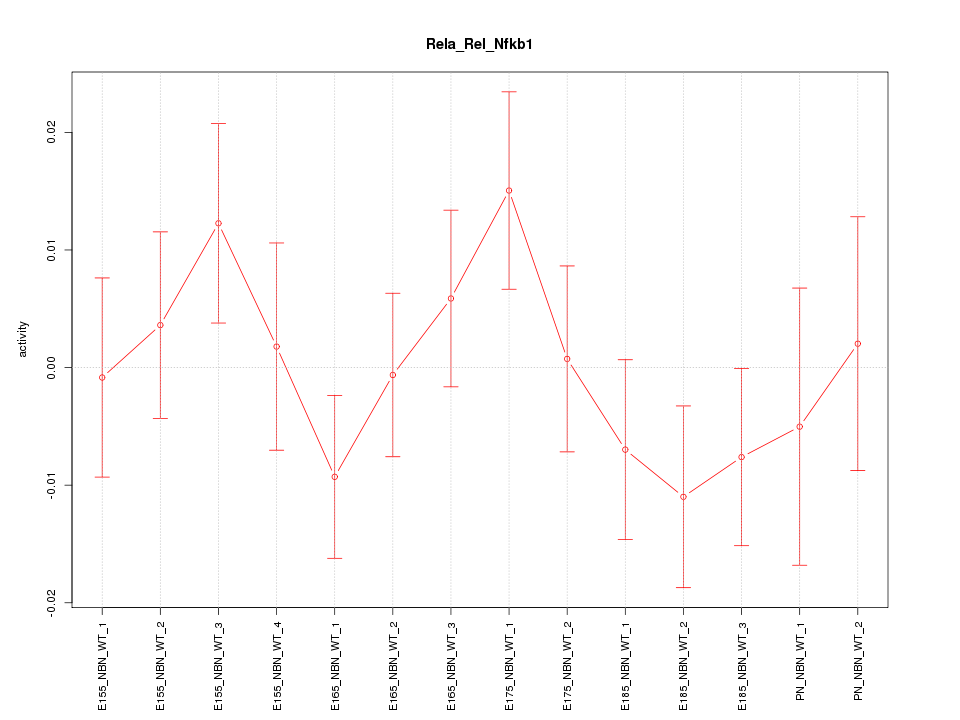
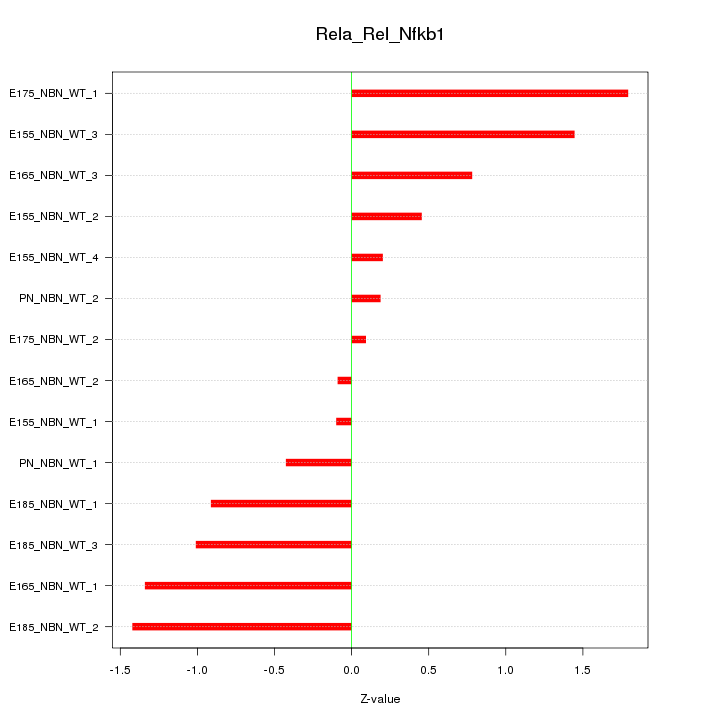
| 0.6 |
1.9 |
GO:1990034 |
calcium ion export from cell(GO:1990034) |
| 0.5 |
2.4 |
GO:0006680 |
glucosylceramide catabolic process(GO:0006680) negative regulation of filopodium assembly(GO:0051490) |
| 0.4 |
1.1 |
GO:0032916 |
positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 0.2 |
0.9 |
GO:0003365 |
establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.2 |
0.6 |
GO:0061144 |
alveolar secondary septum development(GO:0061144) |
| 0.2 |
0.7 |
GO:2000795 |
negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.2 |
1.5 |
GO:0008063 |
Toll signaling pathway(GO:0008063) |
| 0.2 |
0.5 |
GO:0050883 |
musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.2 |
1.1 |
GO:2001199 |
negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.2 |
0.6 |
GO:0032512 |
regulation of protein phosphatase type 2B activity(GO:0032512) positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.2 |
0.5 |
GO:0070428 |
granuloma formation(GO:0002432) negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) toll-like receptor 5 signaling pathway(GO:0034146) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.1 |
0.4 |
GO:0003349 |
epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.1 |
0.4 |
GO:0030070 |
insulin processing(GO:0030070) |
| 0.1 |
0.4 |
GO:0002925 |
positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) detection of peptidoglycan(GO:0032499) negative regulation of interleukin-18 production(GO:0032701) |
| 0.1 |
0.9 |
GO:0007195 |
adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.1 |
0.3 |
GO:1901509 |
regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.1 |
0.3 |
GO:0001827 |
inner cell mass cell fate commitment(GO:0001827) |
| 0.1 |
0.4 |
GO:0072362 |
regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.1 |
0.3 |
GO:0060023 |
soft palate development(GO:0060023) |
| 0.1 |
0.9 |
GO:0036371 |
protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.1 |
0.8 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 |
0.4 |
GO:0010808 |
positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.1 |
0.4 |
GO:2000189 |
positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 |
0.5 |
GO:0035428 |
hexose transmembrane transport(GO:0035428) |
| 0.1 |
0.3 |
GO:0040009 |
regulation of growth rate(GO:0040009) |
| 0.1 |
0.7 |
GO:0002755 |
MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.1 |
0.3 |
GO:0051643 |
endoplasmic reticulum localization(GO:0051643) |
| 0.1 |
0.2 |
GO:0070103 |
regulation of interleukin-6-mediated signaling pathway(GO:0070103) negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 0.1 |
0.7 |
GO:0061088 |
sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 |
0.4 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |
| 0.1 |
0.3 |
GO:0021564 |
glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 0.1 |
0.9 |
GO:1901898 |
negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 |
0.2 |
GO:0042663 |
regulation of endodermal cell fate specification(GO:0042663) |
| 0.1 |
0.4 |
GO:0090383 |
phagosome acidification(GO:0090383) |
| 0.1 |
0.3 |
GO:0035616 |
histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 |
0.4 |
GO:0006957 |
complement activation, alternative pathway(GO:0006957) |
| 0.1 |
1.2 |
GO:0071378 |
growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 |
0.2 |
GO:1901491 |
negative regulation of lymphangiogenesis(GO:1901491) |
| 0.1 |
0.1 |
GO:0045763 |
negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.1 |
0.3 |
GO:0071883 |
activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.1 |
0.2 |
GO:0061300 |
cerebellum vasculature development(GO:0061300) |
| 0.1 |
0.2 |
GO:0030321 |
transepithelial chloride transport(GO:0030321) transepithelial ammonium transport(GO:0070634) |
| 0.1 |
0.2 |
GO:1901731 |
calcium-mediated signaling using extracellular calcium source(GO:0035585) positive regulation of platelet aggregation(GO:1901731) |
| 0.1 |
0.2 |
GO:0043379 |
memory T cell differentiation(GO:0043379) |
| 0.1 |
0.4 |
GO:0007296 |
vitellogenesis(GO:0007296) |
| 0.1 |
0.4 |
GO:0090074 |
negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 |
0.2 |
GO:2000211 |
regulation of glutamate metabolic process(GO:2000211) |
| 0.0 |
0.9 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 |
0.2 |
GO:0032289 |
central nervous system myelin formation(GO:0032289) cardiac cell fate specification(GO:0060912) |
| 0.0 |
0.1 |
GO:0036015 |
response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.0 |
0.2 |
GO:0051708 |
intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) cytosol to ER transport(GO:0046967) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 |
0.1 |
GO:0043201 |
response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.0 |
0.3 |
GO:0002553 |
histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.0 |
0.5 |
GO:1901621 |
negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 |
0.1 |
GO:0021577 |
hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.0 |
0.5 |
GO:0090244 |
Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.0 |
0.3 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.0 |
0.4 |
GO:0061470 |
T follicular helper cell differentiation(GO:0061470) |
| 0.0 |
0.2 |
GO:0044268 |
multicellular organismal protein metabolic process(GO:0044268) |
| 0.0 |
0.0 |
GO:0030050 |
vesicle transport along actin filament(GO:0030050) |
| 0.0 |
0.2 |
GO:0060158 |
phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 |
0.2 |
GO:1902031 |
regulation of NADP metabolic process(GO:1902031) |
| 0.0 |
0.2 |
GO:2000346 |
negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 |
0.4 |
GO:0051574 |
positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 |
0.1 |
GO:0045204 |
MAPK export from nucleus(GO:0045204) |
| 0.0 |
1.1 |
GO:0007212 |
dopamine receptor signaling pathway(GO:0007212) |
| 0.0 |
0.1 |
GO:0097051 |
establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.0 |
0.1 |
GO:1904884 |
telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.0 |
0.2 |
GO:0006646 |
phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 |
0.2 |
GO:0043369 |
CD4-positive or CD8-positive, alpha-beta T cell lineage commitment(GO:0043369) |
| 0.0 |
0.2 |
GO:0045876 |
positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 |
0.2 |
GO:1902373 |
negative regulation of mRNA catabolic process(GO:1902373) |
| 0.0 |
0.2 |
GO:1902035 |
positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 |
0.0 |
GO:0032621 |
interleukin-18 production(GO:0032621) |
| 0.0 |
0.1 |
GO:0071929 |
alpha-tubulin acetylation(GO:0071929) |
| 0.0 |
0.1 |
GO:0021943 |
formation of radial glial scaffolds(GO:0021943) |
| 0.0 |
0.1 |
GO:0070213 |
protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 |
0.1 |
GO:2000110 |
protein sialylation(GO:1990743) negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.0 |
0.3 |
GO:0071225 |
cellular response to muramyl dipeptide(GO:0071225) |
| 0.0 |
0.1 |
GO:0030259 |
lipid glycosylation(GO:0030259) |
| 0.0 |
0.1 |
GO:0021905 |
pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) positive regulation of cell fate specification(GO:0042660) |
| 0.0 |
0.1 |
GO:0006657 |
CDP-choline pathway(GO:0006657) |
| 0.0 |
0.1 |
GO:1902528 |
regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.0 |
0.6 |
GO:0000413 |
protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 |
0.1 |
GO:0060481 |
lobar bronchus epithelium development(GO:0060481) |
| 0.0 |
0.3 |
GO:0046541 |
saliva secretion(GO:0046541) |
| 0.0 |
0.2 |
GO:0075522 |
IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 |
0.3 |
GO:0000185 |
activation of MAPKKK activity(GO:0000185) |
| 0.0 |
0.1 |
GO:0006481 |
C-terminal protein methylation(GO:0006481) |
| 0.0 |
0.1 |
GO:0044314 |
protein K27-linked ubiquitination(GO:0044314) |
| 0.0 |
0.1 |
GO:0006553 |
lysine metabolic process(GO:0006553) |
| 0.0 |
0.1 |
GO:2001168 |
regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 |
0.2 |
GO:0002072 |
optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 |
0.0 |
GO:1901295 |
canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of heart induction(GO:0090381) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of heart induction(GO:1901321) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.0 |
0.2 |
GO:0000022 |
mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 |
0.2 |
GO:0048149 |
behavioral response to ethanol(GO:0048149) |
| 0.0 |
0.1 |
GO:0033136 |
serine phosphorylation of STAT3 protein(GO:0033136) |
| 0.0 |
0.1 |
GO:0032382 |
positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) positive regulation of cholesterol import(GO:1904109) positive regulation of sterol import(GO:2000911) |
| 0.0 |
0.1 |
GO:0046898 |
response to cycloheximide(GO:0046898) |
| 0.0 |
0.1 |
GO:0009597 |
detection of virus(GO:0009597) |
| 0.0 |
0.2 |
GO:0031642 |
negative regulation of myelination(GO:0031642) |
| 0.0 |
0.1 |
GO:1900275 |
negative regulation of phospholipase C activity(GO:1900275) |
| 0.0 |
0.2 |
GO:0061302 |
smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 |
0.0 |
GO:0002678 |
positive regulation of chronic inflammatory response(GO:0002678) |
| 0.0 |
0.1 |
GO:0090666 |
scaRNA localization to Cajal body(GO:0090666) |
| 0.0 |
0.2 |
GO:0030320 |
cellular anion homeostasis(GO:0030002) cellular monovalent inorganic anion homeostasis(GO:0030320) |
| 0.0 |
0.4 |
GO:0001967 |
suckling behavior(GO:0001967) |
| 0.0 |
0.1 |
GO:0048341 |
paraxial mesoderm formation(GO:0048341) |
| 0.0 |
0.1 |
GO:0051764 |
actin crosslink formation(GO:0051764) |
| 0.0 |
0.2 |
GO:0060056 |
mammary gland involution(GO:0060056) |
| 0.0 |
0.0 |
GO:1900060 |
negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.0 |
0.1 |
GO:0072553 |
terminal button organization(GO:0072553) |
| 0.0 |
0.1 |
GO:0038128 |
ERBB2 signaling pathway(GO:0038128) |
| 0.0 |
0.2 |
GO:0090331 |
negative regulation of platelet aggregation(GO:0090331) |
| 0.0 |
0.0 |
GO:1902474 |
positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 |
0.0 |
GO:1903599 |
positive regulation of mitophagy(GO:1903599) |
| 0.0 |
0.1 |
GO:0046726 |
positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 |
0.2 |
GO:0010818 |
T cell chemotaxis(GO:0010818) |
| 0.0 |
0.1 |
GO:0006398 |
mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 |
0.2 |
GO:1902187 |
negative regulation of viral release from host cell(GO:1902187) |
| 0.0 |
0.4 |
GO:0070979 |
protein K11-linked ubiquitination(GO:0070979) |
| 0.0 |
0.1 |
GO:0097341 |
inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 |
0.4 |
GO:0046854 |
phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 |
0.1 |
GO:0072610 |
interleukin-12 secretion(GO:0072610) regulation of interleukin-12 secretion(GO:2001182) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 |
0.9 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 |
0.1 |
GO:1902414 |
protein localization to cell junction(GO:1902414) |
| 0.0 |
0.1 |
GO:0016266 |
O-glycan processing(GO:0016266) |
| 0.0 |
0.1 |
GO:0006547 |
histidine metabolic process(GO:0006547) |
| 0.0 |
0.2 |
GO:0032793 |
positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 |
0.1 |
GO:1904425 |
negative regulation of GTP binding(GO:1904425) |
| 0.0 |
0.0 |
GO:0055099 |
response to high density lipoprotein particle(GO:0055099) |
| 0.0 |
0.1 |
GO:0016322 |
neuron remodeling(GO:0016322) |
| 0.0 |
0.1 |
GO:0032020 |
ISG15-protein conjugation(GO:0032020) |
| 0.0 |
0.2 |
GO:0006903 |
vesicle targeting(GO:0006903) |
| 0.0 |
0.1 |
GO:0060575 |
intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 |
0.6 |
GO:0072661 |
protein targeting to plasma membrane(GO:0072661) |
| 0.0 |
0.3 |
GO:0021522 |
spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 |
0.0 |
GO:0060558 |
regulation of calcidiol 1-monooxygenase activity(GO:0060558) |
| 0.0 |
0.4 |
GO:1900026 |
positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 |
0.0 |
GO:0017183 |
peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 |
0.3 |
GO:0043551 |
regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 |
0.0 |
GO:0045162 |
clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 |
0.3 |
GO:0048488 |
synaptic vesicle endocytosis(GO:0048488) |
| 0.0 |
0.0 |
GO:0002834 |
immune response to tumor cell(GO:0002418) regulation of response to tumor cell(GO:0002834) regulation of immune response to tumor cell(GO:0002837) |
| 0.0 |
0.4 |
GO:0045599 |
negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 |
0.1 |
GO:0010724 |
regulation of definitive erythrocyte differentiation(GO:0010724) |