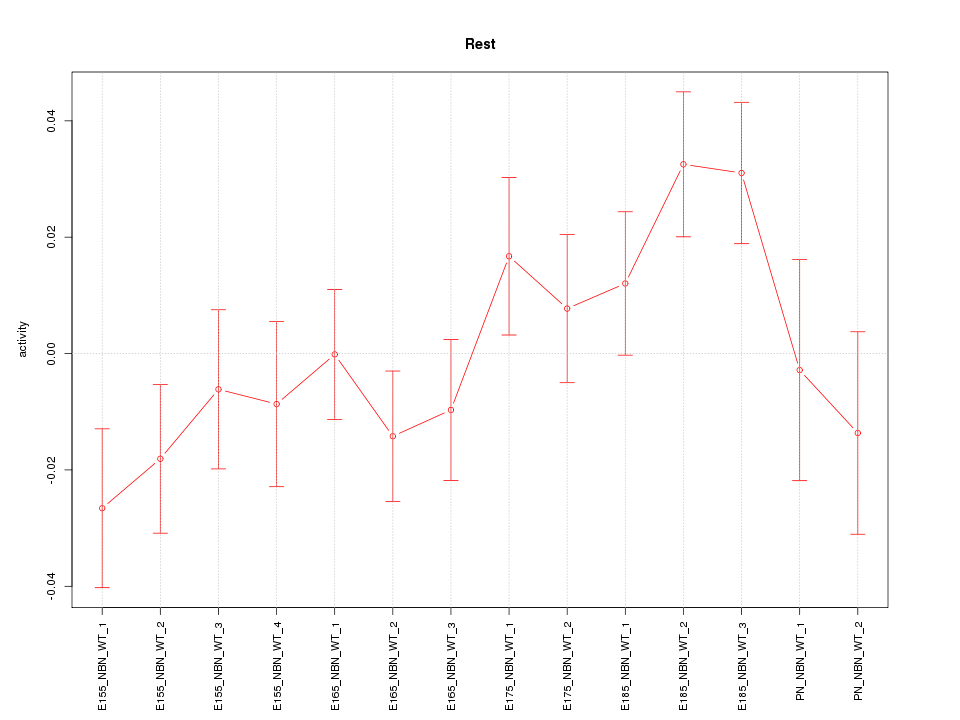
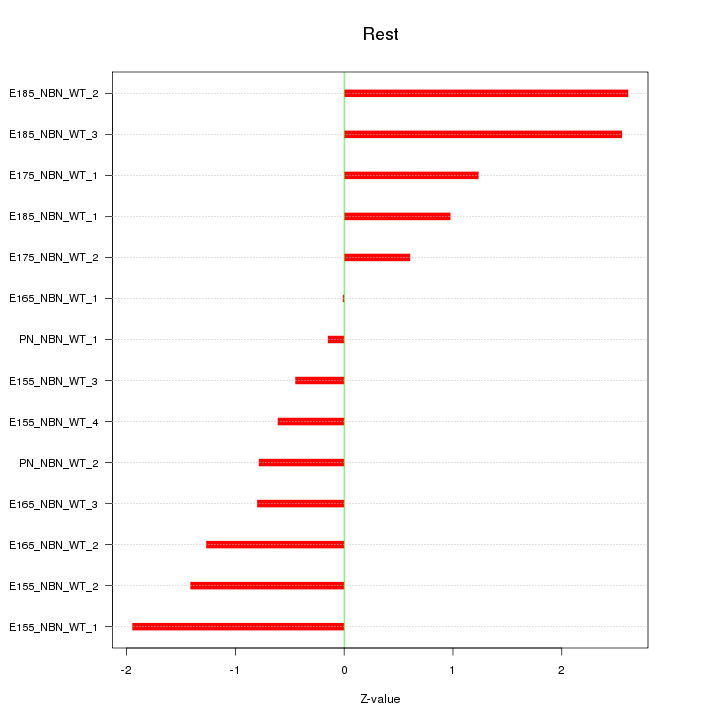
Motif ID: Rest
Z-value: 1.348
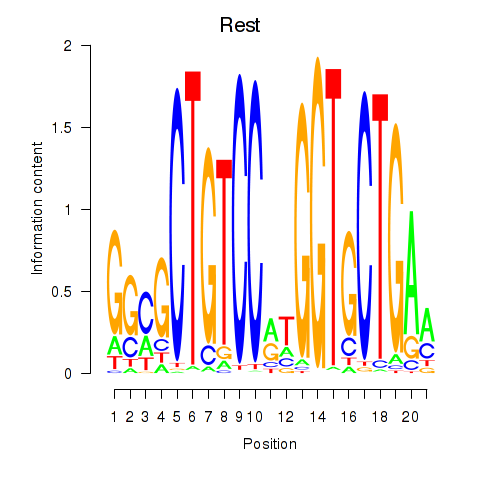
Transcription factors associated with Rest:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Rest | ENSMUSG00000029249.9 | Rest |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rest | mm10_v2_chr5_+_77265454_77265549 | -0.40 | 1.6e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 4.0 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.8 | 2.3 | GO:1900673 | olefin metabolic process(GO:1900673) |
| 0.4 | 1.3 | GO:0030827 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.3 | 1.0 | GO:0006295 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) positive regulation of t-circle formation(GO:1904431) |
| 0.3 | 4.6 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.3 | 0.9 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.3 | 2.0 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.3 | 3.4 | GO:0043084 | penile erection(GO:0043084) |
| 0.2 | 0.9 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.2 | 1.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.2 | 0.9 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.2 | 5.0 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.2 | 0.5 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.2 | 0.5 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.2 | 0.8 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.1 | 1.3 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.1 | 0.3 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.1 | 2.1 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.1 | 0.8 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 | 2.0 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.8 | GO:0001504 | neurotransmitter uptake(GO:0001504) |
| 0.1 | 0.8 | GO:0044331 | cell-cell adhesion mediated by cadherin(GO:0044331) |
| 0.1 | 0.2 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.1 | 0.7 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 0.5 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 | 1.3 | GO:0051894 | integrin activation(GO:0033622) positive regulation of focal adhesion assembly(GO:0051894) |
| 0.1 | 1.1 | GO:0007614 | short-term memory(GO:0007614) |
| 0.0 | 0.3 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 1.3 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 0.5 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.6 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 1.1 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.3 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 | 0.2 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 | 0.2 | GO:1903861 | regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.3 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.3 | 1.0 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) ERCC4-ERCC1 complex(GO:0070522) |
| 0.2 | 1.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 1.3 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.1 | 5.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.8 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 2.9 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 0.2 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 0.1 | 3.4 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 0.3 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 0.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 2.1 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.5 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 1.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.4 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 4.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 2.0 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.1 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.0 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.2 | 2.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.2 | 2.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 3.0 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 1.0 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.1 | 0.9 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.1 | 0.5 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 5.0 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 0.3 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.1 | 1.8 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.1 | 2.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 4.6 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
| 0.1 | 0.9 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 2.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 1.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 2.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.3 | GO:0015375 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 2.2 | GO:0005496 | steroid binding(GO:0005496) |
| 0.0 | 0.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.5 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.8 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.0 | GO:0030249 | cyclase regulator activity(GO:0010851) guanylate cyclase regulator activity(GO:0030249) |
| 0.0 | 0.8 | GO:0042054 | histone methyltransferase activity(GO:0042054) |