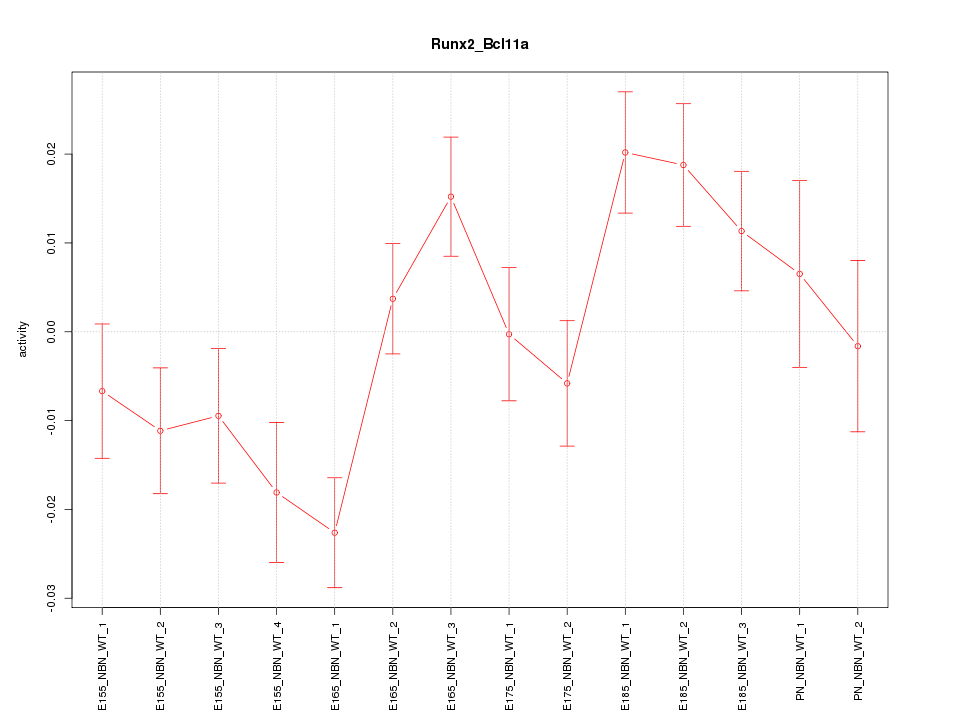
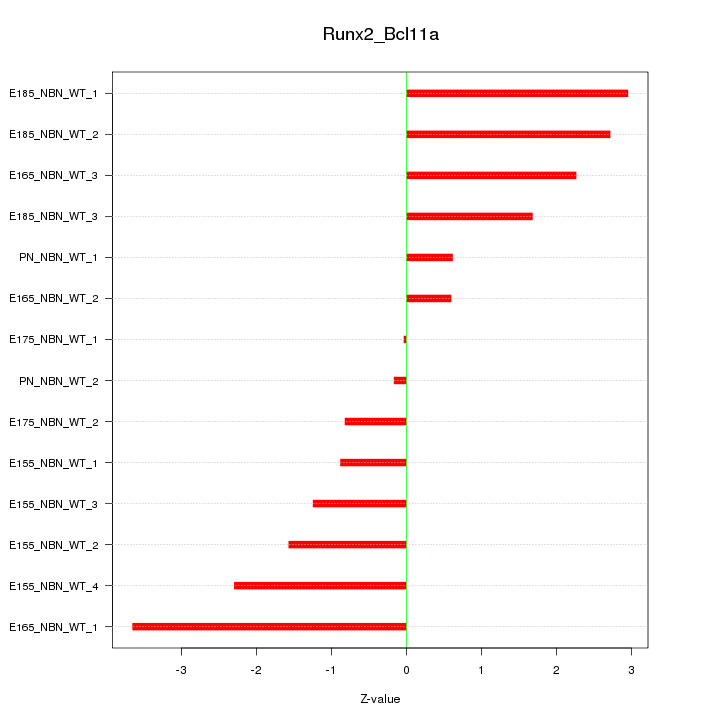
Motif ID: Runx2_Bcl11a
Z-value: 1.872
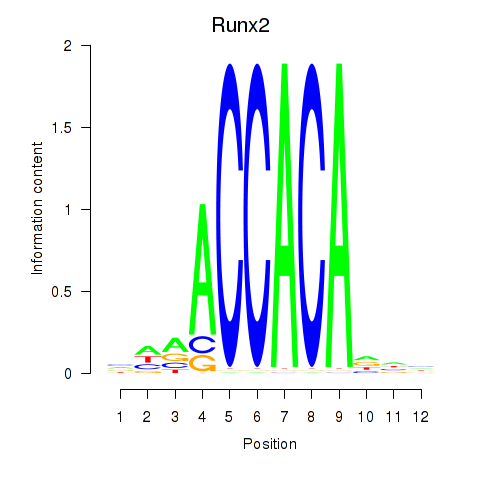
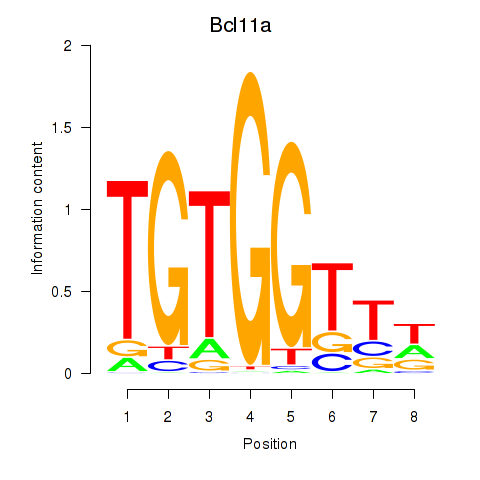
Transcription factors associated with Runx2_Bcl11a:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Bcl11a | ENSMUSG00000000861.9 | Bcl11a |
| Runx2 | ENSMUSG00000039153.10 | Runx2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Bcl11a | mm10_v2_chr11_+_24078173_24078219 | -0.29 | 3.1e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.3 | GO:0034162 | toll-like receptor 9 signaling pathway(GO:0034162) |
| 1.2 | 3.6 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 1.0 | 5.9 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.9 | 2.7 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.8 | 2.5 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.8 | 5.8 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.8 | 7.0 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.8 | 3.9 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) negative regulation of cytolysis(GO:0045918) |
| 0.7 | 2.2 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.6 | 6.9 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.6 | 1.8 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.6 | 1.8 | GO:0036292 | positive regulation of prostaglandin biosynthetic process(GO:0031394) DNA rewinding(GO:0036292) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.5 | 2.7 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.5 | 1.5 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.5 | 0.5 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.5 | 3.8 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.4 | 1.3 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.4 | 1.6 | GO:2001274 | negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.4 | 4.4 | GO:0051923 | sulfation(GO:0051923) |
| 0.4 | 3.0 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.4 | 1.8 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.4 | 0.7 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.3 | 2.4 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.3 | 1.0 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.3 | 1.4 | GO:0010813 | neuropeptide catabolic process(GO:0010813) |
| 0.3 | 1.0 | GO:0032829 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) |
| 0.3 | 2.4 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.3 | 1.9 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.3 | 1.9 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.3 | 2.1 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.3 | 2.9 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.3 | 0.9 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.3 | 5.1 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.3 | 1.9 | GO:0051461 | regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.3 | 0.3 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.3 | 0.5 | GO:0010752 | regulation of cGMP-mediated signaling(GO:0010752) |
| 0.3 | 0.8 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.3 | 0.5 | GO:0045342 | MHC class II biosynthetic process(GO:0045342) |
| 0.3 | 2.5 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.2 | 2.0 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.2 | 1.0 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 0.2 | 0.7 | GO:0021837 | motogenic signaling involved in postnatal olfactory bulb interneuron migration(GO:0021837) positive regulation of mitotic cell cycle DNA replication(GO:1903465) |
| 0.2 | 1.2 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.2 | 0.9 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) positive regulation of oocyte development(GO:0060282) |
| 0.2 | 0.6 | GO:0001803 | antibody-dependent cellular cytotoxicity(GO:0001788) type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 0.2 | 2.4 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.2 | 1.0 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.2 | 0.6 | GO:0045014 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.2 | 0.4 | GO:2000501 | natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.2 | 0.7 | GO:1904177 | regulation of adipose tissue development(GO:1904177) |
| 0.2 | 0.7 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.2 | 0.6 | GO:0030300 | regulation of intestinal cholesterol absorption(GO:0030300) |
| 0.2 | 1.3 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.2 | 0.9 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.2 | 1.0 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.2 | 0.8 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.2 | 0.5 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.2 | 1.2 | GO:0015862 | uridine transport(GO:0015862) |
| 0.2 | 0.7 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.2 | 1.0 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.2 | 0.5 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.2 | 0.3 | GO:0045938 | positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.2 | 0.6 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.2 | 0.2 | GO:1903463 | regulation of mitotic cell cycle DNA replication(GO:1903463) |
| 0.1 | 0.4 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.1 | 0.7 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.1 | 0.4 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.1 | 0.6 | GO:0032229 | negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.1 | 0.8 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 | 1.4 | GO:0033280 | response to vitamin D(GO:0033280) |
| 0.1 | 0.4 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) terminal web assembly(GO:1902896) |
| 0.1 | 1.1 | GO:0071493 | cellular response to UV-B(GO:0071493) replicative senescence(GO:0090399) |
| 0.1 | 0.2 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 0.7 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 | 0.4 | GO:0035622 | intrahepatic bile duct development(GO:0035622) cholangiocyte proliferation(GO:1990705) |
| 0.1 | 0.8 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 0.2 | GO:0002034 | angiotensin mediated vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001998) regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 0.1 | 0.7 | GO:0045988 | negative regulation of striated muscle contraction(GO:0045988) |
| 0.1 | 0.5 | GO:2000587 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.1 | 2.2 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 | 0.5 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.1 | 1.2 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.1 | 0.3 | GO:0010957 | negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.1 | 0.7 | GO:0097460 | ferrous iron import into cell(GO:0097460) |
| 0.1 | 1.2 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.1 | 0.5 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 1.0 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.1 | 0.3 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.1 | 1.4 | GO:0021766 | hippocampus development(GO:0021766) |
| 0.1 | 0.4 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.1 | 0.3 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.1 | 0.5 | GO:0043312 | neutrophil degranulation(GO:0043312) |
| 0.1 | 0.5 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 0.9 | GO:0046036 | GTP biosynthetic process(GO:0006183) CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.1 | 0.2 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.1 | 0.6 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 0.5 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.1 | 0.3 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.1 | 0.3 | GO:1900533 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.1 | 0.5 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.1 | 0.6 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.1 | 0.4 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.1 | 0.5 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.1 | 0.2 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 0.5 | GO:1904667 | negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.1 | 0.2 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 0.2 | GO:0071873 | response to norepinephrine(GO:0071873) |
| 0.1 | 0.4 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.1 | 0.6 | GO:0021546 | rhombomere development(GO:0021546) |
| 0.1 | 0.2 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.1 | 0.5 | GO:0021894 | cerebral cortex GABAergic interneuron development(GO:0021894) |
| 0.1 | 0.3 | GO:2001015 | negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.1 | 0.3 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.1 | 0.2 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.1 | 0.3 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.1 | 2.2 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 1.6 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.1 | 0.3 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.1 | 0.5 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 0.2 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.2 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 0.3 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.1 | 0.5 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 1.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.2 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 0.2 | GO:0006842 | tricarboxylic acid transport(GO:0006842) succinate transport(GO:0015744) citrate transport(GO:0015746) |
| 0.1 | 0.2 | GO:1900145 | regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900175) positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.1 | 0.7 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.3 | GO:1904721 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.1 | 1.5 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.1 | 0.1 | GO:0001996 | regulation of systemic arterial blood pressure by norepinephrine-epinephrine(GO:0001993) positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) |
| 0.1 | 1.2 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 1.1 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 0.2 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 1.0 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.7 | GO:0071357 | type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.3 | GO:0060055 | angiogenesis involved in wound healing(GO:0060055) |
| 0.0 | 1.0 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.2 | GO:0060978 | angiogenesis involved in coronary vascular morphogenesis(GO:0060978) |
| 0.0 | 0.2 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.2 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.3 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 1.0 | GO:0014741 | negative regulation of muscle hypertrophy(GO:0014741) |
| 0.0 | 0.2 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.3 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.0 | 0.3 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.7 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.9 | GO:0007614 | short-term memory(GO:0007614) |
| 0.0 | 0.2 | GO:2001270 | regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) |
| 0.0 | 0.5 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 0.3 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.0 | 0.3 | GO:2000252 | negative regulation of feeding behavior(GO:2000252) |
| 0.0 | 0.2 | GO:0010917 | negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.0 | 0.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:2000279 | negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.0 | 0.2 | GO:1902514 | positive regulation of Rac protein signal transduction(GO:0035022) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 0.1 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.3 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.2 | GO:0032811 | negative regulation of epinephrine secretion(GO:0032811) |
| 0.0 | 0.3 | GO:0042711 | maternal behavior(GO:0042711) |
| 0.0 | 0.2 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.0 | 0.4 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.4 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.4 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.1 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.0 | 0.4 | GO:0090201 | negative regulation of release of cytochrome c from mitochondria(GO:0090201) |
| 0.0 | 0.1 | GO:0046013 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.2 | GO:0046502 | uroporphyrinogen III metabolic process(GO:0046502) |
| 0.0 | 0.3 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.7 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.3 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.4 | GO:0045909 | positive regulation of vasodilation(GO:0045909) |
| 0.0 | 0.3 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.2 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.8 | GO:0048662 | negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.0 | 0.3 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.1 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.1 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 1.7 | GO:0050810 | regulation of steroid biosynthetic process(GO:0050810) |
| 0.0 | 0.1 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.0 | 0.1 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.2 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.2 | GO:1903427 | negative regulation of reactive oxygen species biosynthetic process(GO:1903427) |
| 0.0 | 0.1 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.1 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.1 | GO:0098501 | polynucleotide dephosphorylation(GO:0098501) |
| 0.0 | 0.1 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.0 | 0.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.4 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.2 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.2 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.0 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.0 | 0.1 | GO:1903894 | regulation of IRE1-mediated unfolded protein response(GO:1903894) |
| 0.0 | 0.2 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.2 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.3 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.7 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.1 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.0 | 0.2 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.0 | 0.1 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 1.3 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.0 | 0.1 | GO:0061718 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.7 | GO:1902808 | positive regulation of cell cycle G1/S phase transition(GO:1902808) |
| 0.0 | 0.2 | GO:0036506 | maintenance of unfolded protein(GO:0036506) protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 | 2.3 | GO:0048515 | spermatid differentiation(GO:0048515) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.3 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.4 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.1 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:0055089 | fatty acid homeostasis(GO:0055089) |
| 0.0 | 0.1 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.1 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.1 | GO:0021940 | positive regulation of cerebellar granule cell precursor proliferation(GO:0021940) |
| 0.0 | 0.1 | GO:0015819 | lysine transport(GO:0015819) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.1 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.1 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.2 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 0.0 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) |
| 0.0 | 0.2 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.3 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.2 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.2 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.2 | GO:0008209 | androgen metabolic process(GO:0008209) |
| 0.0 | 0.1 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.1 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) establishment of protein localization to mitochondrial membrane(GO:0090151) |
| 0.0 | 0.5 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.2 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.1 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.1 | GO:0032911 | nerve growth factor production(GO:0032902) negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 | 0.0 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.2 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.0 | 0.3 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.1 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.4 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.2 | GO:0044253 | positive regulation of collagen metabolic process(GO:0010714) positive regulation of multicellular organismal metabolic process(GO:0044253) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.1 | GO:0060161 | peptidyl-arginine omega-N-methylation(GO:0035247) histone H4-R3 methylation(GO:0043985) positive regulation of dopamine receptor signaling pathway(GO:0060161) |
| 0.0 | 1.0 | GO:0051592 | response to calcium ion(GO:0051592) |
| 0.0 | 0.4 | GO:0019915 | lipid storage(GO:0019915) |
| 0.0 | 0.7 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.2 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.1 | GO:0051788 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 6.4 | GO:0032009 | early phagosome(GO:0032009) |
| 0.4 | 1.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.4 | 3.5 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.4 | 4.6 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.4 | 1.1 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.4 | 1.5 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.3 | 1.2 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.3 | 0.9 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.3 | 2.5 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.3 | 0.8 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.2 | 1.7 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.2 | 0.7 | GO:0044299 | C-fiber(GO:0044299) |
| 0.2 | 1.8 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.2 | 0.8 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.2 | 16.8 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.2 | 0.5 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.2 | 0.5 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.1 | 2.6 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 0.6 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.1 | 0.7 | GO:0043256 | laminin complex(GO:0043256) |
| 0.1 | 1.6 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 1.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 1.5 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.7 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 2.5 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.5 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.1 | 1.6 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 1.0 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 0.3 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 0.1 | 0.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 2.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.8 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 0.3 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 0.4 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 0.4 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 0.4 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 0.2 | GO:0038045 | large latent transforming growth factor-beta complex(GO:0038045) |
| 0.1 | 0.2 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.1 | 0.7 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.1 | 0.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.9 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 0.5 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.1 | 0.6 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 0.3 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 0.7 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.1 | 0.5 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 0.7 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.4 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.1 | 2.9 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.1 | 0.2 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 1.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.2 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.3 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.2 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.6 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.2 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.2 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 1.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.3 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.3 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.1 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.0 | 0.5 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.8 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 1.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 24.8 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.2 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.1 | GO:1990462 | omegasome(GO:1990462) |
| 0.0 | 0.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.3 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 1.2 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.5 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.2 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.7 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 1.0 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 2.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.2 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.3 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.1 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 2.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.0 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 1.6 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.2 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.0 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.0 | 0.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 0.7 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.6 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.2 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.7 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.2 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.1 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.7 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.3 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.2 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.1 | GO:0071439 | clathrin complex(GO:0071439) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 5.9 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 1.5 | 4.4 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 1.3 | 3.9 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 1.0 | 7.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 1.0 | 4.0 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.9 | 3.6 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.7 | 2.7 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.6 | 2.5 | GO:0070573 | tripeptidyl-peptidase activity(GO:0008240) metallodipeptidase activity(GO:0070573) |
| 0.5 | 4.8 | GO:0035325 | Toll-like receptor binding(GO:0035325) |
| 0.5 | 1.9 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.4 | 1.3 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.4 | 1.8 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) phospholipase A2 inhibitor activity(GO:0019834) |
| 0.4 | 2.5 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.4 | 1.2 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) arginine binding(GO:0034618) |
| 0.4 | 1.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.4 | 1.5 | GO:0031721 | haptoglobin binding(GO:0031720) hemoglobin alpha binding(GO:0031721) |
| 0.4 | 8.2 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.3 | 2.8 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.3 | 1.4 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.3 | 1.4 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.3 | 3.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.3 | 1.8 | GO:0043426 | MRF binding(GO:0043426) |
| 0.3 | 2.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.3 | 1.0 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.2 | 1.2 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.2 | 2.7 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.2 | 0.7 | GO:0031755 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.2 | 0.9 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.2 | 0.6 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.2 | 0.5 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.2 | 3.8 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.2 | 2.4 | GO:0008430 | selenium binding(GO:0008430) |
| 0.2 | 0.5 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.2 | 0.6 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.2 | 0.5 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 2.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.5 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.1 | 0.9 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.1 | 0.5 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 2.8 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.7 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.4 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 0.7 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 0.4 | GO:0005118 | sevenless binding(GO:0005118) |
| 0.1 | 2.1 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 0.2 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.1 | 1.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.4 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.1 | 1.6 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 0.6 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 0.7 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.1 | 0.6 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.8 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 1.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.8 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.6 | GO:0016453 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.1 | 0.3 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.1 | 1.1 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.1 | 0.3 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.1 | 0.7 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.1 | 0.8 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 1.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.7 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.5 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.1 | 0.5 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.5 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 1.0 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 0.3 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.2 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.1 | 0.9 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 0.4 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.8 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.5 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.1 | 0.2 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.1 | 0.3 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.2 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.1 | 1.1 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 1.4 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.4 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.1 | 0.7 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 1.0 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 0.5 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.3 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.1 | 0.1 | GO:0004935 | adrenergic receptor activity(GO:0004935) alpha-adrenergic receptor activity(GO:0004936) |
| 0.1 | 0.2 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.0 | 0.7 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.2 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.5 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.0 | 0.7 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.4 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 1.0 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.3 | GO:0031802 | G-protein coupled adenosine receptor activity(GO:0001609) type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.9 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.8 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.2 | GO:0098634 | protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.3 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.7 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.3 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.4 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.4 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.4 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.8 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.2 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.1 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.0 | 0.1 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) |
| 0.0 | 0.4 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.3 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 1.4 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 1.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 1.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.3 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.2 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) |
| 0.0 | 0.2 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.2 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 1.0 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.3 | GO:0034943 | acyl-CoA ligase activity(GO:0003996) 3-oxo-2-(2'-pentenyl)cyclopentane-1-octanoic acid CoA ligase activity(GO:0010435) 3-isopropenyl-6-oxoheptanoyl-CoA synthetase activity(GO:0018854) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA synthetase activity(GO:0018855) benzoyl acetate-CoA ligase activity(GO:0018856) 2,4-dichlorobenzoate-CoA ligase activity(GO:0018857) very long-chain fatty acid-CoA ligase activity(GO:0031957) pivalate-CoA ligase activity(GO:0034783) cyclopropanecarboxylate-CoA ligase activity(GO:0034793) adipate-CoA ligase activity(GO:0034796) citronellyl-CoA ligase activity(GO:0034823) mentha-1,3-dione-CoA ligase activity(GO:0034841) thiophene-2-carboxylate-CoA ligase activity(GO:0034842) 2,4,4-trimethylpentanoate-CoA ligase activity(GO:0034865) cis-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034942) trans-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034943) branched-chain acyl-CoA synthetase (ADP-forming) activity(GO:0043759) aryl-CoA synthetase (ADP-forming) activity(GO:0043762) 3-hydroxypropionyl-CoA synthetase activity(GO:0043955) perillic acid:CoA ligase (ADP-forming) activity(GO:0052685) perillic acid:CoA ligase (AMP-forming) activity(GO:0052686) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (ADP-forming) activity(GO:0052687) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (AMP-forming) activity(GO:0052688) pristanate-CoA ligase activity(GO:0070251) malonyl-CoA synthetase activity(GO:0090409) |
| 0.0 | 0.3 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.2 | GO:0045703 | pinocarveol dehydrogenase activity(GO:0018446) chloral hydrate dehydrogenase activity(GO:0018447) hydroxymethylmethylsilanediol oxidase activity(GO:0018448) 1-phenylethanol dehydrogenase activity(GO:0018449) myrtenol dehydrogenase activity(GO:0018450) cis-1,2-dihydroxy-1,2-dihydro-8-carboxynaphthalene dehydrogenase activity(GO:0034522) 3-hydroxy-4-methyloctanoyl-CoA dehydrogenase activity(GO:0034582) 2-hydroxy-4-isopropenylcyclohexane-1-carboxyl-CoA dehydrogenase activity(GO:0034778) cis-9,10-dihydroanthracene-9,10-diol dehydrogenase activity(GO:0034817) citronellol dehydrogenase activity(GO:0034821) naphthyl-2-hydroxymethyl-succinyl-CoA dehydrogenase activity(GO:0034847) 2,4,4-trimethyl-1-pentanol dehydrogenase activity(GO:0034863) 2,4,4-trimethyl-3-hydroxypentanoyl-CoA dehydrogenase activity(GO:0034868) 1-hydroxy-4,4-dimethylpentan-3-one dehydrogenase activity(GO:0034871) endosulfan diol dehydrogenase activity(GO:0034891) endosulfan hydroxyether dehydrogenase activity(GO:0034901) 3-hydroxy-2-methylhexanoyl-CoA dehydrogenase activity(GO:0034918) 3-hydroxy-2,6-dimethyl-5-methylene-heptanoyl-CoA dehydrogenase activity(GO:0034944) versicolorin reductase activity(GO:0042469) ketoreductase activity(GO:0045703) |
| 0.0 | 0.2 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 0.2 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.0 | 0.6 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 2.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.2 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.0 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.0 | 0.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.8 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.0 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.0 | 0.0 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 0.2 | GO:0070636 | single-strand selective uracil DNA N-glycosylase activity(GO:0017065) nicotinamide riboside hydrolase activity(GO:0070635) nicotinic acid riboside hydrolase activity(GO:0070636) deoxyribonucleoside 5'-monophosphate N-glycosidase activity(GO:0070694) |
| 0.0 | 0.1 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 1.4 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.2 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.3 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.1 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.2 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 1.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.6 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.3 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.1 | GO:0046403 | polynucleotide 3'-phosphatase activity(GO:0046403) |
| 0.0 | 0.6 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.3 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.0 | 0.1 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.2 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.1 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.1 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0016917 | GABA receptor activity(GO:0016917) |
| 0.0 | 0.0 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.0 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.0 | 5.0 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.0 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.6 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 0.3 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.1 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.0 | 0.1 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |