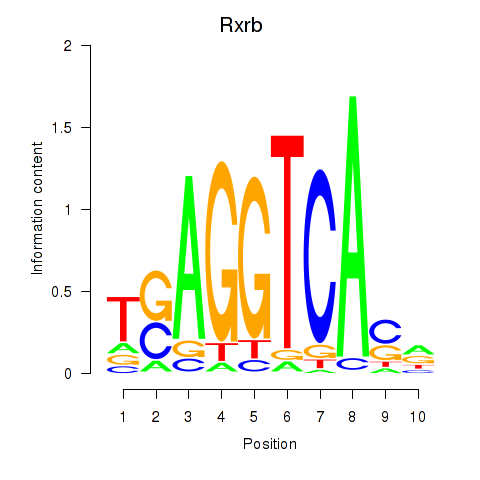
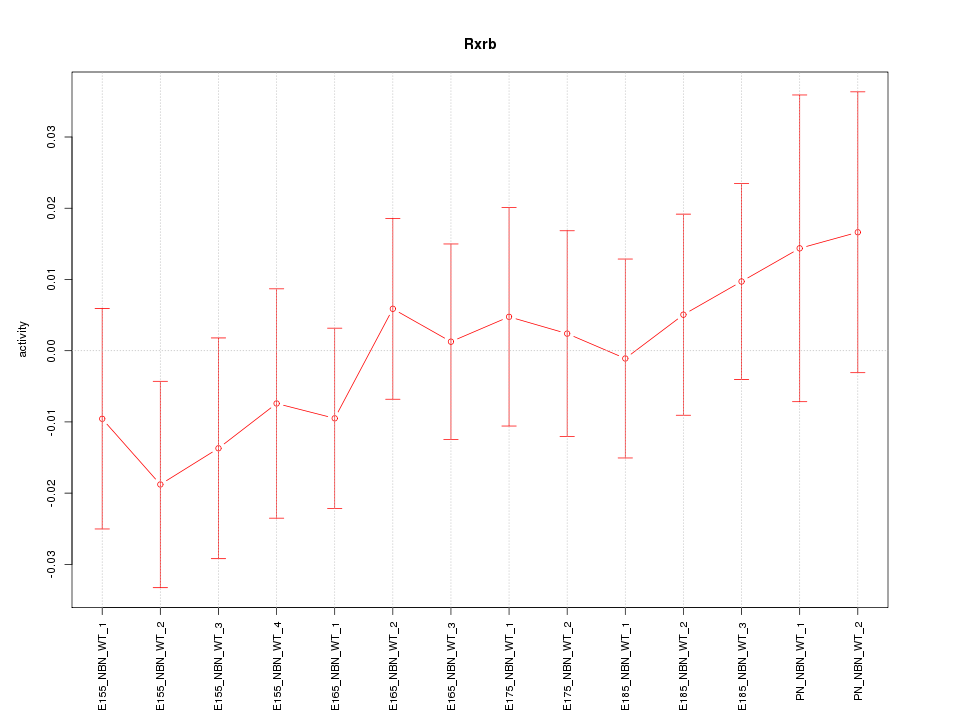
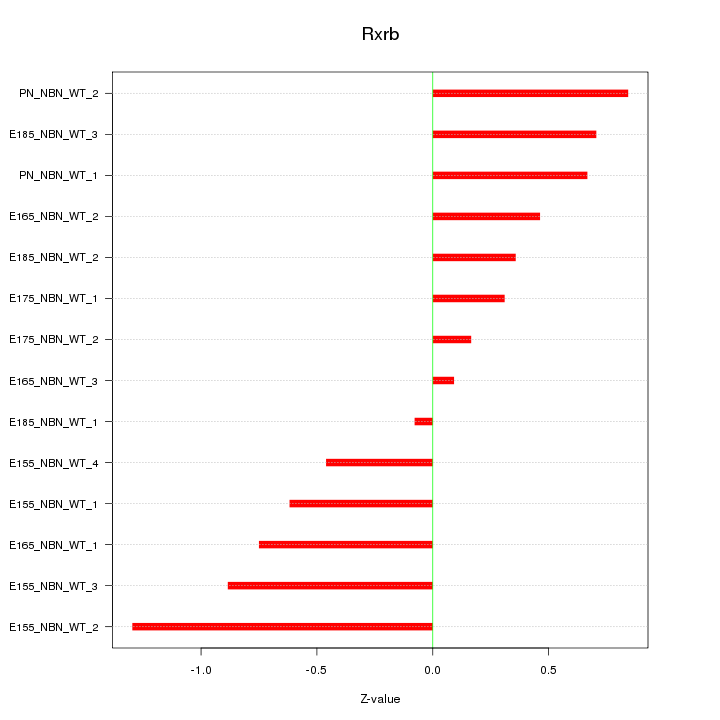
| 0.5 |
3.3 |
GO:0006957 |
complement activation, alternative pathway(GO:0006957) |
| 0.3 |
0.9 |
GO:0050929 |
corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.2 |
0.6 |
GO:1903795 |
regulation of inorganic anion transmembrane transport(GO:1903795) |
| 0.2 |
0.7 |
GO:1903463 |
mitotic cell cycle phase(GO:0098763) regulation of mitotic cell cycle DNA replication(GO:1903463) |
| 0.2 |
0.8 |
GO:0030951 |
establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 |
0.4 |
GO:0060854 |
patterning of lymph vessels(GO:0060854) |
| 0.1 |
1.7 |
GO:0010388 |
cullin deneddylation(GO:0010388) |
| 0.1 |
0.4 |
GO:0019372 |
lipoxygenase pathway(GO:0019372) |
| 0.1 |
0.6 |
GO:0034316 |
negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 |
0.7 |
GO:2000042 |
negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 |
0.3 |
GO:0015910 |
peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 |
0.3 |
GO:0097494 |
regulation of vesicle size(GO:0097494) |
| 0.1 |
0.4 |
GO:2001269 |
positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.1 |
0.3 |
GO:0051890 |
regulation of cardioblast differentiation(GO:0051890) |
| 0.1 |
0.7 |
GO:0051764 |
actin crosslink formation(GO:0051764) |
| 0.0 |
0.1 |
GO:1900365 |
positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 |
0.8 |
GO:0090136 |
epithelial cell-cell adhesion(GO:0090136) |
| 0.0 |
1.2 |
GO:0045662 |
negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 |
0.3 |
GO:0010571 |
positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 |
0.2 |
GO:1903525 |
regulation of membrane tubulation(GO:1903525) |
| 0.0 |
0.9 |
GO:0010614 |
negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 |
0.6 |
GO:0007190 |
activation of adenylate cyclase activity(GO:0007190) |
| 0.0 |
0.1 |
GO:0097623 |
potassium ion export across plasma membrane(GO:0097623) |
| 0.0 |
0.8 |
GO:0000028 |
ribosomal small subunit assembly(GO:0000028) |
| 0.0 |
0.3 |
GO:2000348 |
regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 |
0.3 |
GO:0006855 |
drug transmembrane transport(GO:0006855) |
| 0.0 |
0.8 |
GO:0070208 |
protein heterotrimerization(GO:0070208) |
| 0.0 |
1.1 |
GO:0031146 |
SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 |
0.2 |
GO:0034393 |
positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 |
0.1 |
GO:0021847 |
ventricular zone neuroblast division(GO:0021847) |
| 0.0 |
0.7 |
GO:0045742 |
positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) |
| 0.0 |
0.4 |
GO:0046548 |
retinal rod cell development(GO:0046548) |
| 0.0 |
0.1 |
GO:0072008 |
glomerular mesangial cell differentiation(GO:0072008) glomerular mesangial cell development(GO:0072144) |
| 0.0 |
0.8 |
GO:1901522 |
positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 |
0.3 |
GO:0051894 |
positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 |
0.1 |
GO:0014719 |
skeletal muscle satellite cell activation(GO:0014719) |
| 0.0 |
1.5 |
GO:0043149 |
contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 |
0.2 |
GO:0010225 |
response to UV-C(GO:0010225) |
| 0.0 |
0.1 |
GO:0015868 |
purine ribonucleotide transport(GO:0015868) |
| 0.0 |
0.2 |
GO:0045624 |
positive regulation of T-helper cell differentiation(GO:0045624) |