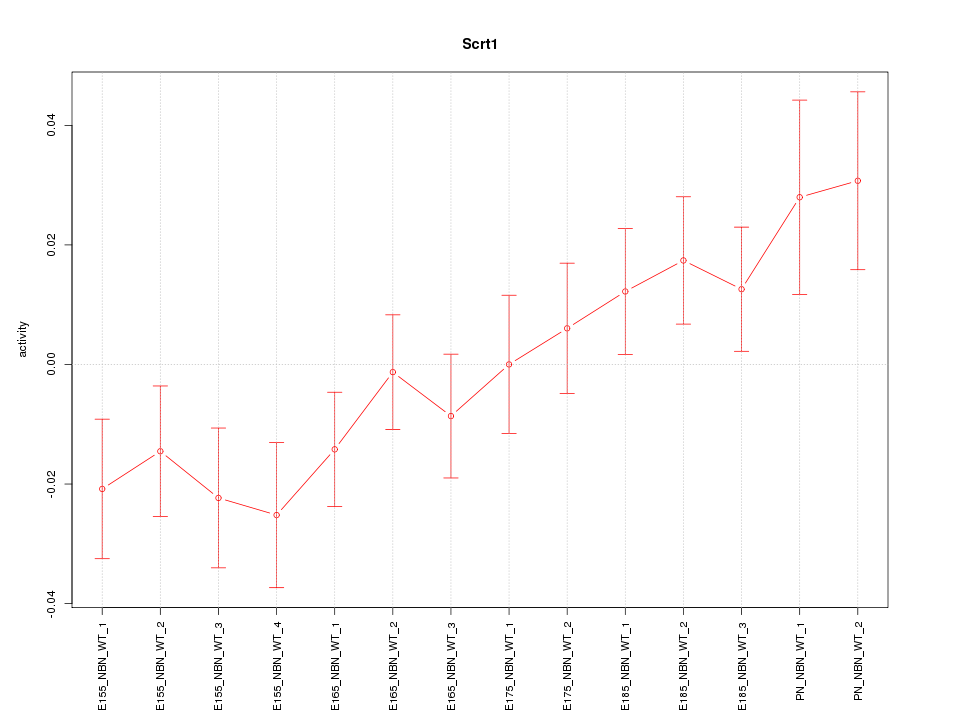
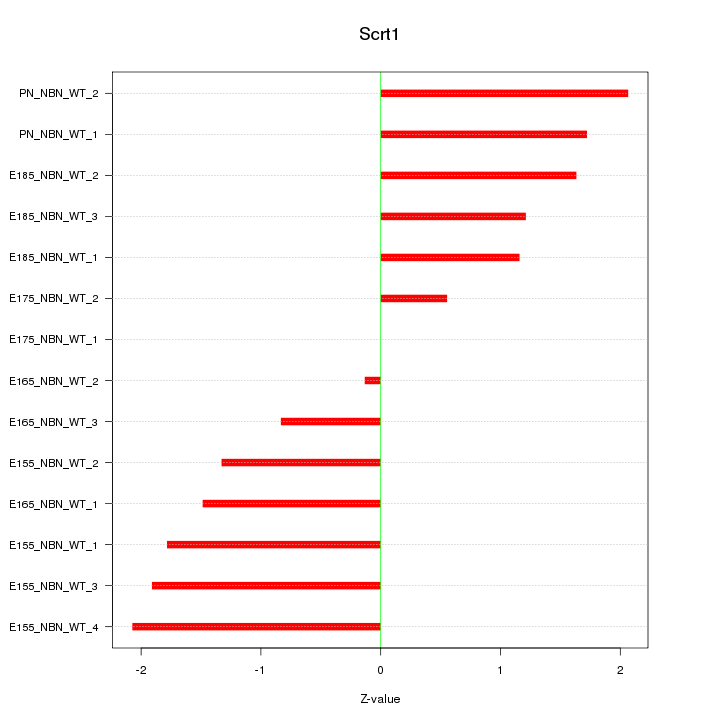
Motif ID: Scrt1
Z-value: 1.435
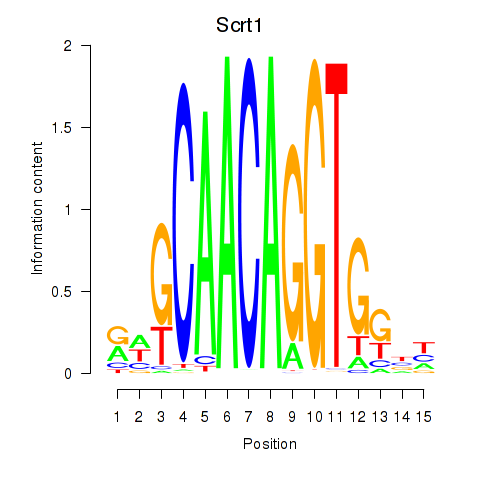
Transcription factors associated with Scrt1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Scrt1 | ENSMUSG00000048385.8 | Scrt1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Scrt1 | mm10_v2_chr15_-_76521902_76522129 | -0.93 | 1.6e-06 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.2 | GO:0042939 | renal sodium ion transport(GO:0003096) glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 1.6 | 4.7 | GO:0072034 | renal vesicle induction(GO:0072034) |
| 1.3 | 4.0 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 1.2 | 3.6 | GO:0051464 | positive regulation of cortisol secretion(GO:0051464) |
| 1.2 | 4.8 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.7 | 3.7 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.6 | 2.5 | GO:0032901 | positive regulation of neurotrophin production(GO:0032901) |
| 0.6 | 1.8 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.6 | 2.4 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.5 | 1.5 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.4 | 1.8 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.4 | 1.2 | GO:2000564 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) CD8-positive, alpha-beta T cell proliferation(GO:0035740) negative regulation of regulatory T cell differentiation(GO:0045590) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.4 | 1.1 | GO:0015920 | regulation of phosphatidylcholine catabolic process(GO:0010899) lipopolysaccharide transport(GO:0015920) |
| 0.4 | 2.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.4 | 1.4 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.3 | 2.4 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.3 | 2.3 | GO:0071699 | endodermal cell fate specification(GO:0001714) olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.3 | 1.6 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.3 | 4.8 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.3 | 3.1 | GO:0003093 | regulation of glomerular filtration(GO:0003093) |
| 0.3 | 1.4 | GO:1902309 | positive regulation of mast cell chemotaxis(GO:0060754) negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.3 | 1.7 | GO:0003383 | apical constriction(GO:0003383) |
| 0.2 | 0.7 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.2 | 2.4 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.2 | 1.1 | GO:0021508 | floor plate formation(GO:0021508) floor plate morphogenesis(GO:0033505) |
| 0.2 | 2.7 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.2 | 2.3 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.2 | 0.8 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.2 | 1.4 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.2 | 5.7 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.2 | 1.6 | GO:0045056 | transcytosis(GO:0045056) |
| 0.2 | 1.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 2.5 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 1.0 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 0.9 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 3.0 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 0.6 | GO:0015744 | succinate transport(GO:0015744) |
| 0.1 | 4.5 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.1 | 1.1 | GO:0016114 | terpenoid biosynthetic process(GO:0016114) |
| 0.1 | 0.4 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 1.1 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.6 | GO:0042891 | antibiotic transport(GO:0042891) |
| 0.1 | 0.4 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.1 | 1.2 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 1.1 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 0.5 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.1 | 3.2 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.1 | 3.0 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.1 | 4.1 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 3.0 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.1 | 2.4 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.1 | 1.7 | GO:0034142 | toll-like receptor 4 signaling pathway(GO:0034142) |
| 0.1 | 0.2 | GO:0039530 | MDA-5 signaling pathway(GO:0039530) |
| 0.1 | 0.2 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.1 | 4.1 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.1 | 0.3 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 0.2 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 | 0.3 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 | 0.5 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.1 | 0.3 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.1 | 1.4 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.1 | 0.1 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.1 | 0.5 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 0.3 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 1.0 | GO:0009072 | aromatic amino acid family metabolic process(GO:0009072) |
| 0.1 | 0.9 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 0.3 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.1 | 1.9 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 0.2 | GO:0071033 | nuclear retention of pre-mRNA at the site of transcription(GO:0071033) CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.3 | GO:0045583 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.0 | 0.2 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.6 | GO:1903861 | regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 1.1 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.4 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) |
| 0.0 | 0.4 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 3.6 | GO:0051592 | response to calcium ion(GO:0051592) |
| 0.0 | 0.2 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.0 | 0.9 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 1.4 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.0 | 0.2 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.0 | 0.4 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 1.7 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.5 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 0.4 | GO:0017144 | drug metabolic process(GO:0017144) |
| 0.0 | 0.3 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.2 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.0 | 0.1 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 | 0.1 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.9 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.0 | 0.4 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.7 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.4 | GO:0030201 | heparan sulfate proteoglycan metabolic process(GO:0030201) |
| 0.0 | 0.2 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.0 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.0 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.6 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.8 | 2.4 | GO:0000811 | GINS complex(GO:0000811) |
| 0.8 | 3.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.7 | 4.0 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.4 | 1.4 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.3 | 1.0 | GO:0036501 | UFD1-NPL4 complex(GO:0036501) |
| 0.3 | 1.7 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.2 | 1.7 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.2 | 0.9 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 3.0 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 0.4 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 1.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 1.5 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.1 | 0.9 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 1.1 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 0.3 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 1.1 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 0.2 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.1 | 2.2 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.1 | 0.5 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 0.4 | GO:0044613 | nuclear pore central transport channel(GO:0044613) Flemming body(GO:0090543) |
| 0.1 | 1.4 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 5.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 0.5 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 2.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.7 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 2.5 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 5.0 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 1.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 2.1 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 3.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.7 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.2 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.1 | GO:0019908 | nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 3.3 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 2.7 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 1.0 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.6 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 2.4 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.8 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 4.2 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.1 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 1.2 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 13.3 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 4.2 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.3 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 1.6 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 1.0 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.3 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 0.5 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 2.5 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.4 | GO:0035869 | ciliary transition zone(GO:0035869) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.6 | 5.2 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.4 | 1.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.4 | 4.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.3 | 1.0 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.3 | 2.5 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.3 | 2.4 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.3 | 1.1 | GO:0004337 | geranyltranstransferase activity(GO:0004337) |
| 0.3 | 1.3 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.3 | 4.0 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.2 | 2.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.2 | 0.6 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.2 | 0.8 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.2 | 1.0 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.2 | 1.4 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 2.4 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.1 | 1.5 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 0.6 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 0.1 | 1.9 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 0.6 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 0.1 | 1.7 | GO:0005536 | glucose binding(GO:0005536) |
| 0.1 | 1.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 1.7 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 0.3 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 1.9 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 0.5 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 1.8 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.4 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 7.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 5.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.4 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 0.3 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.1 | 0.3 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 1.0 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.1 | 0.9 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 2.5 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.3 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 0.3 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 3.0 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 0.2 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 1.0 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.9 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 8.6 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 1.7 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.3 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 1.6 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.4 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 1.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.9 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 1.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 2.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.1 | GO:0098631 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.2 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 4.3 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.7 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.3 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.0 | 2.7 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 1.1 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.3 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 1.1 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 7.0 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.4 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.4 | GO:0030276 | clathrin binding(GO:0030276) |