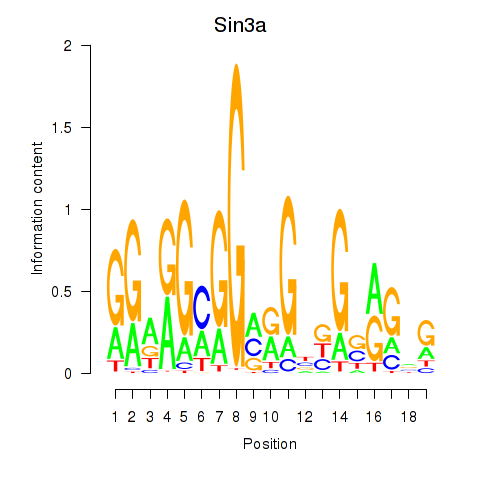
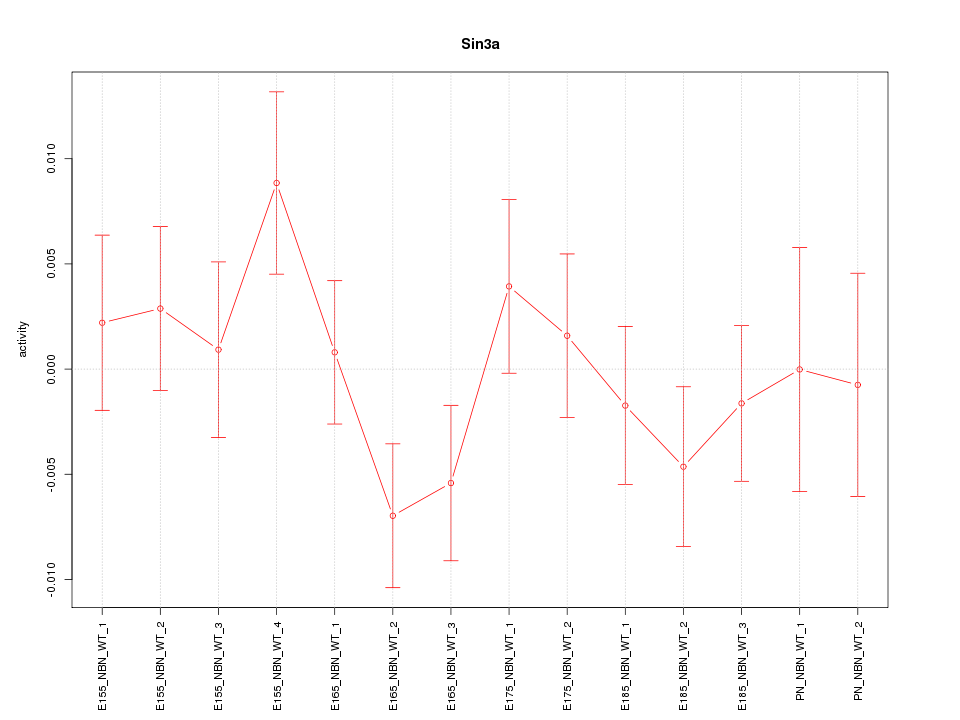
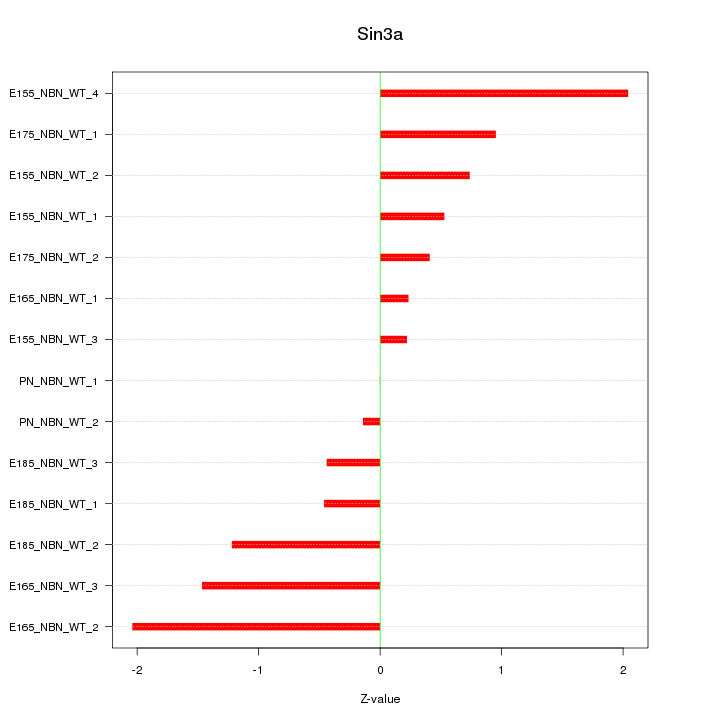
| 0.7 |
2.2 |
GO:0050975 |
sensory perception of touch(GO:0050975) |
| 0.4 |
2.2 |
GO:0021965 |
spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.4 |
1.6 |
GO:0051643 |
endoplasmic reticulum localization(GO:0051643) |
| 0.3 |
1.0 |
GO:0032240 |
negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.3 |
1.0 |
GO:1902528 |
regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.3 |
1.0 |
GO:0003221 |
right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.3 |
0.9 |
GO:0060244 |
negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.3 |
1.1 |
GO:2000795 |
negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.3 |
0.8 |
GO:0016191 |
synaptic vesicle uncoating(GO:0016191) regulation of endosome organization(GO:1904978) positive regulation of endosome organization(GO:1904980) |
| 0.3 |
0.8 |
GO:0032847 |
regulation of cellular pH reduction(GO:0032847) |
| 0.3 |
2.1 |
GO:0051775 |
response to redox state(GO:0051775) |
| 0.3 |
2.0 |
GO:0006477 |
protein sulfation(GO:0006477) |
| 0.3 |
0.8 |
GO:0003349 |
epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.2 |
2.4 |
GO:0061470 |
T follicular helper cell differentiation(GO:0061470) |
| 0.2 |
0.2 |
GO:0045974 |
miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.2 |
0.7 |
GO:1901254 |
positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.2 |
1.2 |
GO:2000301 |
negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.2 |
0.7 |
GO:0060399 |
positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.2 |
0.6 |
GO:1903244 |
positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.2 |
0.6 |
GO:0002331 |
pre-B cell allelic exclusion(GO:0002331) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.2 |
0.8 |
GO:2000211 |
regulation of glutamate metabolic process(GO:2000211) |
| 0.2 |
1.1 |
GO:0030643 |
cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.2 |
0.5 |
GO:0032915 |
positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.2 |
1.0 |
GO:0070863 |
positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.2 |
1.6 |
GO:0048386 |
positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.2 |
0.8 |
GO:2000774 |
positive regulation of cellular senescence(GO:2000774) |
| 0.2 |
0.6 |
GO:0042276 |
error-prone translesion synthesis(GO:0042276) |
| 0.1 |
0.6 |
GO:0090283 |
regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.1 |
0.4 |
GO:0050925 |
negative regulation of negative chemotaxis(GO:0050925) |
| 0.1 |
0.6 |
GO:2001032 |
regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.1 |
1.1 |
GO:0097118 |
neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.1 |
1.1 |
GO:0050882 |
voluntary musculoskeletal movement(GO:0050882) |
| 0.1 |
0.3 |
GO:2000286 |
receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.1 |
0.6 |
GO:0090234 |
regulation of kinetochore assembly(GO:0090234) |
| 0.1 |
0.4 |
GO:0060489 |
establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.1 |
0.1 |
GO:0042440 |
pigment metabolic process(GO:0042440) |
| 0.1 |
0.5 |
GO:0021698 |
cerebellar cortex structural organization(GO:0021698) |
| 0.1 |
0.6 |
GO:0060158 |
phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 |
0.4 |
GO:0007181 |
transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.1 |
1.2 |
GO:0048681 |
negative regulation of axon regeneration(GO:0048681) |
| 0.1 |
0.4 |
GO:0048208 |
vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 |
0.5 |
GO:0043974 |
histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.1 |
0.1 |
GO:0003219 |
cardiac right ventricle formation(GO:0003219) |
| 0.1 |
0.3 |
GO:0042713 |
sperm ejaculation(GO:0042713) |
| 0.1 |
0.8 |
GO:0017196 |
N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 |
0.4 |
GO:0002069 |
columnar/cuboidal epithelial cell maturation(GO:0002069) |
| 0.1 |
0.3 |
GO:0035864 |
response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.1 |
1.2 |
GO:0021785 |
branchiomotor neuron axon guidance(GO:0021785) |
| 0.1 |
0.3 |
GO:0060596 |
mammary placode formation(GO:0060596) |
| 0.1 |
0.1 |
GO:0070103 |
regulation of interleukin-6-mediated signaling pathway(GO:0070103) negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 0.1 |
0.6 |
GO:1900246 |
positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.1 |
0.5 |
GO:1901843 |
positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.1 |
0.2 |
GO:0072053 |
renal inner medulla development(GO:0072053) renal outer medulla development(GO:0072054) |
| 0.1 |
0.3 |
GO:2000616 |
negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.1 |
0.3 |
GO:0032058 |
positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.1 |
0.5 |
GO:0060447 |
bud outgrowth involved in lung branching(GO:0060447) |
| 0.1 |
0.2 |
GO:1901668 |
regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 |
0.3 |
GO:0008228 |
opsonization(GO:0008228) |
| 0.1 |
0.8 |
GO:0006620 |
posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 |
0.3 |
GO:0010871 |
negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.1 |
0.2 |
GO:0043988 |
histone H3-S28 phosphorylation(GO:0043988) |
| 0.1 |
0.6 |
GO:0045925 |
positive regulation of female receptivity(GO:0045925) |
| 0.1 |
0.6 |
GO:0097368 |
establishment of Sertoli cell barrier(GO:0097368) |
| 0.1 |
0.9 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 |
0.6 |
GO:0044829 |
positive regulation by host of viral genome replication(GO:0044829) |
| 0.1 |
0.7 |
GO:0006265 |
DNA topological change(GO:0006265) |
| 0.1 |
0.3 |
GO:0002184 |
cytoplasmic translational termination(GO:0002184) |
| 0.1 |
0.4 |
GO:0048631 |
regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.1 |
0.4 |
GO:0032439 |
endosome localization(GO:0032439) |
| 0.1 |
0.9 |
GO:0051967 |
negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 |
1.0 |
GO:0006857 |
oligopeptide transport(GO:0006857) |
| 0.1 |
0.3 |
GO:0033326 |
cerebrospinal fluid secretion(GO:0033326) |
| 0.1 |
1.2 |
GO:1990403 |
embryonic brain development(GO:1990403) |
| 0.1 |
0.7 |
GO:0043353 |
enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 |
0.2 |
GO:0097688 |
AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.1 |
1.0 |
GO:0001711 |
endodermal cell fate commitment(GO:0001711) |
| 0.1 |
1.3 |
GO:0060081 |
membrane hyperpolarization(GO:0060081) |
| 0.1 |
0.5 |
GO:0000056 |
ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 |
0.3 |
GO:0061010 |
gall bladder development(GO:0061010) |
| 0.1 |
0.4 |
GO:0034436 |
glycoprotein transport(GO:0034436) |
| 0.1 |
0.4 |
GO:0001842 |
neural fold formation(GO:0001842) |
| 0.1 |
0.4 |
GO:0002182 |
cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.1 |
0.2 |
GO:0018076 |
N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.1 |
0.5 |
GO:0010747 |
positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.1 |
0.2 |
GO:0051695 |
actin filament uncapping(GO:0051695) |
| 0.1 |
0.2 |
GO:2000224 |
testosterone biosynthetic process(GO:0061370) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.1 |
0.8 |
GO:0035970 |
peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 |
0.2 |
GO:0048352 |
neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) |
| 0.1 |
0.8 |
GO:0010650 |
positive regulation of cell communication by electrical coupling(GO:0010650) cellular response to magnesium ion(GO:0071286) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 |
0.7 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 0.1 |
0.5 |
GO:0000022 |
mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.1 |
0.6 |
GO:0071420 |
cellular response to histamine(GO:0071420) |
| 0.1 |
0.1 |
GO:0071896 |
protein localization to adherens junction(GO:0071896) |
| 0.1 |
0.3 |
GO:0090168 |
Golgi reassembly(GO:0090168) |
| 0.1 |
0.2 |
GO:1903237 |
negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 |
0.3 |
GO:0045759 |
negative regulation of action potential(GO:0045759) |
| 0.1 |
0.5 |
GO:0060164 |
regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 |
0.5 |
GO:0060903 |
positive regulation of meiosis I(GO:0060903) |
| 0.1 |
0.1 |
GO:0030950 |
establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 |
0.4 |
GO:0051409 |
response to nitrosative stress(GO:0051409) |
| 0.1 |
0.6 |
GO:1901660 |
calcium ion export(GO:1901660) |
| 0.1 |
0.4 |
GO:0031297 |
replication fork processing(GO:0031297) DNA-dependent DNA replication maintenance of fidelity(GO:0045005) |
| 0.1 |
0.2 |
GO:0045110 |
intermediate filament bundle assembly(GO:0045110) |
| 0.1 |
0.1 |
GO:2000812 |
regulation of barbed-end actin filament capping(GO:2000812) |
| 0.1 |
0.2 |
GO:0034145 |
positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.1 |
0.1 |
GO:0060693 |
regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.1 |
0.2 |
GO:2000304 |
positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.1 |
0.4 |
GO:2000973 |
regulation of pro-B cell differentiation(GO:2000973) |
| 0.1 |
0.4 |
GO:0061158 |
3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 |
0.2 |
GO:0006667 |
sphinganine metabolic process(GO:0006667) |
| 0.1 |
0.4 |
GO:0061343 |
cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.1 |
0.2 |
GO:0016256 |
N-glycan processing to lysosome(GO:0016256) |
| 0.1 |
0.2 |
GO:0055071 |
cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.1 |
0.2 |
GO:0035616 |
histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 |
0.4 |
GO:0036072 |
intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 |
0.1 |
GO:0097116 |
gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.1 |
0.3 |
GO:1900122 |
positive regulation of receptor binding(GO:1900122) |
| 0.1 |
0.5 |
GO:0090394 |
negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 0.1 |
0.5 |
GO:0045634 |
regulation of melanocyte differentiation(GO:0045634) |
| 0.1 |
0.3 |
GO:0043267 |
negative regulation of potassium ion transport(GO:0043267) |
| 0.1 |
0.3 |
GO:0070562 |
regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 |
0.2 |
GO:0042998 |
positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.1 |
0.7 |
GO:0060013 |
righting reflex(GO:0060013) |
| 0.1 |
1.3 |
GO:0008045 |
motor neuron axon guidance(GO:0008045) |
| 0.1 |
0.6 |
GO:0060159 |
regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.1 |
0.2 |
GO:0048211 |
Golgi vesicle docking(GO:0048211) |
| 0.1 |
0.2 |
GO:1903215 |
negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.1 |
0.2 |
GO:0071692 |
protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) |
| 0.1 |
0.3 |
GO:0019336 |
phenol-containing compound catabolic process(GO:0019336) |
| 0.1 |
0.2 |
GO:0040009 |
nucleolus to nucleoplasm transport(GO:0032066) regulation of growth rate(GO:0040009) |
| 0.1 |
0.3 |
GO:0071894 |
histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.1 |
0.2 |
GO:0046460 |
triglyceride biosynthetic process(GO:0019432) neutral lipid biosynthetic process(GO:0046460) acylglycerol biosynthetic process(GO:0046463) |
| 0.1 |
0.3 |
GO:0071699 |
olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.1 |
0.6 |
GO:0051481 |
negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.1 |
0.1 |
GO:2000744 |
anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.0 |
0.4 |
GO:0035563 |
positive regulation of chromatin binding(GO:0035563) |
| 0.0 |
0.3 |
GO:0021747 |
cochlear nucleus development(GO:0021747) |
| 0.0 |
0.4 |
GO:0019065 |
receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 |
0.3 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.0 |
0.1 |
GO:0038026 |
reelin-mediated signaling pathway(GO:0038026) |
| 0.0 |
0.4 |
GO:2000601 |
positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 |
0.1 |
GO:0014042 |
positive regulation of neuron maturation(GO:0014042) |
| 0.0 |
1.0 |
GO:0010971 |
positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.0 |
0.1 |
GO:0060051 |
negative regulation of protein glycosylation(GO:0060051) |
| 0.0 |
0.2 |
GO:1903715 |
regulation of aerobic respiration(GO:1903715) |
| 0.0 |
0.5 |
GO:0030828 |
positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.0 |
0.2 |
GO:0006543 |
glutamine catabolic process(GO:0006543) |
| 0.0 |
0.1 |
GO:0070345 |
negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 |
0.1 |
GO:0006030 |
chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 |
0.2 |
GO:0070425 |
negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 0.0 |
0.3 |
GO:0098532 |
histone H3-K27 trimethylation(GO:0098532) |
| 0.0 |
0.2 |
GO:1903361 |
protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 |
0.3 |
GO:0035507 |
regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 |
0.5 |
GO:0036120 |
cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.0 |
0.3 |
GO:0071044 |
histone mRNA catabolic process(GO:0071044) |
| 0.0 |
0.2 |
GO:0061526 |
acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) acetylcholine secretion(GO:0061526) |
| 0.0 |
0.2 |
GO:2000189 |
positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 |
0.1 |
GO:0097107 |
postsynaptic density organization(GO:0097106) postsynaptic density assembly(GO:0097107) |
| 0.0 |
0.8 |
GO:0045653 |
negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 |
0.1 |
GO:0060066 |
establishment of endothelial blood-brain barrier(GO:0014045) central nervous system vasculogenesis(GO:0022009) oviduct development(GO:0060066) |
| 0.0 |
0.2 |
GO:0042780 |
tRNA 3'-end processing(GO:0042780) |
| 0.0 |
0.3 |
GO:0034047 |
regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.0 |
0.1 |
GO:0051599 |
response to hydrostatic pressure(GO:0051599) |
| 0.0 |
0.3 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 |
0.2 |
GO:0051660 |
cortical microtubule organization(GO:0043622) establishment of centrosome localization(GO:0051660) |
| 0.0 |
0.2 |
GO:0035372 |
protein localization to microtubule(GO:0035372) |
| 0.0 |
0.3 |
GO:0043568 |
positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 |
0.1 |
GO:0070844 |
misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 |
0.3 |
GO:0000395 |
mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 |
0.2 |
GO:0002924 |
negative regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002924) |
| 0.0 |
0.1 |
GO:0060024 |
rhythmic synaptic transmission(GO:0060024) |
| 0.0 |
0.2 |
GO:1901524 |
regulation of macromitophagy(GO:1901524) negative regulation of macromitophagy(GO:1901525) |
| 0.0 |
0.1 |
GO:2000686 |
regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 |
0.4 |
GO:0016322 |
neuron remodeling(GO:0016322) |
| 0.0 |
0.8 |
GO:0045722 |
positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 |
0.4 |
GO:0061339 |
establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.0 |
0.5 |
GO:0055090 |
acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 |
0.0 |
GO:0061153 |
trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 |
0.7 |
GO:0032148 |
activation of protein kinase B activity(GO:0032148) |
| 0.0 |
0.3 |
GO:0000083 |
regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 |
0.2 |
GO:0051534 |
negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 |
0.1 |
GO:1904742 |
regulation of telomeric DNA binding(GO:1904742) |
| 0.0 |
0.1 |
GO:0014043 |
negative regulation of neuron maturation(GO:0014043) |
| 0.0 |
0.1 |
GO:0006529 |
asparagine biosynthetic process(GO:0006529) |
| 0.0 |
0.1 |
GO:0001779 |
natural killer cell differentiation(GO:0001779) |
| 0.0 |
0.1 |
GO:0097343 |
ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.0 |
0.2 |
GO:1900095 |
regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 |
0.4 |
GO:0043923 |
positive regulation by host of viral transcription(GO:0043923) |
| 0.0 |
0.2 |
GO:0070102 |
interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.0 |
0.2 |
GO:0007089 |
traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 |
0.1 |
GO:0015772 |
disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 |
0.1 |
GO:0086046 |
membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 |
0.4 |
GO:0031936 |
negative regulation of chromatin silencing(GO:0031936) |
| 0.0 |
0.2 |
GO:0031293 |
membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 |
0.1 |
GO:0090148 |
membrane fission(GO:0090148) |
| 0.0 |
0.1 |
GO:0060282 |
positive regulation of oocyte development(GO:0060282) |
| 0.0 |
0.7 |
GO:0060122 |
inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 |
0.4 |
GO:0007252 |
I-kappaB phosphorylation(GO:0007252) |
| 0.0 |
0.3 |
GO:0060052 |
neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 |
0.1 |
GO:0060059 |
embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 |
0.1 |
GO:0045918 |
negative regulation of cytolysis(GO:0045918) |
| 0.0 |
0.2 |
GO:0006983 |
ER overload response(GO:0006983) |
| 0.0 |
0.2 |
GO:0045654 |
positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 |
0.2 |
GO:1900121 |
negative regulation of receptor binding(GO:1900121) |
| 0.0 |
0.4 |
GO:0046855 |
phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 |
0.2 |
GO:0016266 |
O-glycan processing(GO:0016266) |
| 0.0 |
0.2 |
GO:1904707 |
positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.0 |
0.2 |
GO:0060766 |
negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.0 |
0.4 |
GO:0010225 |
response to UV-C(GO:0010225) |
| 0.0 |
0.9 |
GO:0043666 |
regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 |
0.1 |
GO:0045659 |
eosinophil differentiation(GO:0030222) regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.0 |
0.1 |
GO:1900149 |
positive regulation of Schwann cell migration(GO:1900149) |
| 0.0 |
0.1 |
GO:0046654 |
tetrahydrofolate biosynthetic process(GO:0046654) embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 |
0.1 |
GO:0015871 |
choline transport(GO:0015871) |
| 0.0 |
0.1 |
GO:1902277 |
negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.0 |
0.1 |
GO:0010993 |
regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.0 |
0.0 |
GO:0006566 |
threonine metabolic process(GO:0006566) |
| 0.0 |
0.2 |
GO:0098543 |
detection of bacterium(GO:0016045) detection of other organism(GO:0098543) |
| 0.0 |
0.1 |
GO:0070537 |
histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 |
0.3 |
GO:0043928 |
exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 |
0.1 |
GO:0002710 |
negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 |
0.2 |
GO:0030948 |
negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 |
0.1 |
GO:0009405 |
pathogenesis(GO:0009405) |
| 0.0 |
0.1 |
GO:0021691 |
cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 |
0.0 |
GO:0071880 |
adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.0 |
0.6 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.0 |
0.1 |
GO:0002934 |
desmosome organization(GO:0002934) |
| 0.0 |
0.2 |
GO:0006544 |
glycine metabolic process(GO:0006544) |
| 0.0 |
0.1 |
GO:0060915 |
fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) coronal suture morphogenesis(GO:0060365) squamous basal epithelial stem cell differentiation involved in prostate gland acinus development(GO:0060529) fibroblast growth factor receptor signaling pathway involved in mammary gland specification(GO:0060595) mammary gland bud formation(GO:0060615) branch elongation involved in salivary gland morphogenesis(GO:0060667) mesenchymal cell differentiation involved in lung development(GO:0060915) |
| 0.0 |
0.1 |
GO:0018214 |
peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 |
0.1 |
GO:1901642 |
purine nucleoside transmembrane transport(GO:0015860) purine-containing compound transmembrane transport(GO:0072530) nucleoside transmembrane transport(GO:1901642) |
| 0.0 |
0.5 |
GO:0060416 |
response to growth hormone(GO:0060416) |
| 0.0 |
0.6 |
GO:0000413 |
protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 |
0.1 |
GO:2000587 |
regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 |
0.1 |
GO:0032792 |
negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 |
0.2 |
GO:0097369 |
sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 |
0.1 |
GO:0048570 |
notochord morphogenesis(GO:0048570) |
| 0.0 |
0.0 |
GO:0045875 |
negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 |
0.1 |
GO:0021564 |
glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 0.0 |
0.1 |
GO:0060025 |
regulation of synaptic activity(GO:0060025) |
| 0.0 |
0.2 |
GO:0055089 |
fatty acid homeostasis(GO:0055089) |
| 0.0 |
0.3 |
GO:0043162 |
ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 |
0.2 |
GO:0008354 |
germ cell migration(GO:0008354) |
| 0.0 |
0.0 |
GO:0036515 |
serotonergic neuron axon guidance(GO:0036515) |
| 0.0 |
0.1 |
GO:0031915 |
positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 |
0.2 |
GO:0061157 |
mRNA destabilization(GO:0061157) |
| 0.0 |
0.1 |
GO:0097623 |
potassium ion export across plasma membrane(GO:0097623) |
| 0.0 |
0.0 |
GO:0010730 |
negative regulation of hydrogen peroxide metabolic process(GO:0010727) negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.0 |
0.1 |
GO:0023019 |
signal transduction involved in regulation of gene expression(GO:0023019) |
| 0.0 |
0.2 |
GO:0097264 |
self proteolysis(GO:0097264) |
| 0.0 |
0.1 |
GO:0019074 |
viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 |
0.4 |
GO:0007216 |
G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 |
0.4 |
GO:0035024 |
negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 |
0.2 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.0 |
0.1 |
GO:0060510 |
Type II pneumocyte differentiation(GO:0060510) |
| 0.0 |
0.2 |
GO:0050910 |
detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 |
0.0 |
GO:0032509 |
endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 |
0.1 |
GO:1903984 |
regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 |
0.4 |
GO:2000311 |
regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 |
0.3 |
GO:0070884 |
regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 |
0.5 |
GO:0045956 |
positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 |
0.1 |
GO:1902177 |
positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902177) |
| 0.0 |
0.7 |
GO:0097120 |
receptor localization to synapse(GO:0097120) |
| 0.0 |
0.1 |
GO:0006867 |
asparagine transport(GO:0006867) glutamine transport(GO:0006868) histidine transport(GO:0015817) |
| 0.0 |
0.2 |
GO:1904262 |
negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 |
0.3 |
GO:0051292 |
nuclear pore complex assembly(GO:0051292) |
| 0.0 |
0.1 |
GO:0071335 |
hair follicle cell proliferation(GO:0071335) |
| 0.0 |
0.1 |
GO:0070535 |
histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.0 |
0.1 |
GO:0033169 |
histone H3-K9 demethylation(GO:0033169) |
| 0.0 |
0.1 |
GO:0072610 |
interleukin-12 secretion(GO:0072610) regulation of interleukin-12 secretion(GO:2001182) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 |
0.1 |
GO:2001185 |
regulation of CD8-positive, alpha-beta T cell activation(GO:2001185) |
| 0.0 |
0.1 |
GO:2000124 |
regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 |
0.4 |
GO:0010172 |
embryonic body morphogenesis(GO:0010172) |
| 0.0 |
0.1 |
GO:0072674 |
multinuclear osteoclast differentiation(GO:0072674) |
| 0.0 |
0.1 |
GO:0090091 |
positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 |
0.1 |
GO:0036092 |
phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 |
0.2 |
GO:0010457 |
centriole-centriole cohesion(GO:0010457) |
| 0.0 |
0.1 |
GO:0048149 |
behavioral response to ethanol(GO:0048149) |
| 0.0 |
0.0 |
GO:2000821 |
regulation of grooming behavior(GO:2000821) |
| 0.0 |
0.4 |
GO:2000052 |
positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 |
0.1 |
GO:0003430 |
growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.0 |
0.0 |
GO:0035886 |
vascular smooth muscle cell differentiation(GO:0035886) |
| 0.0 |
0.1 |
GO:0048162 |
preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.0 |
0.1 |
GO:0060267 |
positive regulation of respiratory burst(GO:0060267) |
| 0.0 |
0.2 |
GO:0006012 |
galactose metabolic process(GO:0006012) |
| 0.0 |
0.1 |
GO:0034312 |
diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) sphingoid biosynthetic process(GO:0046520) |
| 0.0 |
0.2 |
GO:0006560 |
proline metabolic process(GO:0006560) |
| 0.0 |
0.4 |
GO:0034243 |
regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.0 |
0.1 |
GO:1903897 |
regulation of PERK-mediated unfolded protein response(GO:1903897) |
| 0.0 |
0.1 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.0 |
0.1 |
GO:0072318 |
clathrin coat disassembly(GO:0072318) |
| 0.0 |
0.1 |
GO:0031937 |
positive regulation of chromatin silencing(GO:0031937) |
| 0.0 |
0.3 |
GO:0007064 |
mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 |
0.2 |
GO:0051764 |
actin crosslink formation(GO:0051764) |
| 0.0 |
0.1 |
GO:0060136 |
embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 |
0.4 |
GO:0046854 |
phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 |
0.4 |
GO:0001937 |
negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.0 |
0.1 |
GO:0018242 |
protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 |
0.0 |
GO:0060295 |
regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 |
0.3 |
GO:0006471 |
protein ADP-ribosylation(GO:0006471) |
| 0.0 |
0.2 |
GO:0032876 |
negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 |
0.0 |
GO:0021764 |
amygdala development(GO:0021764) |
| 0.0 |
0.3 |
GO:0045104 |
intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 |
0.0 |
GO:0014063 |
negative regulation of serotonin secretion(GO:0014063) |
| 0.0 |
0.2 |
GO:0070816 |
phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 |
0.1 |
GO:0071257 |
cellular response to electrical stimulus(GO:0071257) |
| 0.0 |
0.1 |
GO:0070933 |
histone H4 deacetylation(GO:0070933) |
| 0.0 |
0.1 |
GO:0001550 |
ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 |
0.0 |
GO:0015793 |
glycerol transport(GO:0015793) |
| 0.0 |
0.2 |
GO:1904152 |
negative regulation of protein exit from endoplasmic reticulum(GO:0070862) regulation of retrograde protein transport, ER to cytosol(GO:1904152) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 |
0.1 |
GO:0008105 |
asymmetric protein localization(GO:0008105) |
| 0.0 |
0.0 |
GO:0098910 |
regulation of atrial cardiac muscle cell action potential(GO:0098910) |
| 0.0 |
0.2 |
GO:0090161 |
Golgi ribbon formation(GO:0090161) |
| 0.0 |
0.2 |
GO:0001672 |
regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 |
0.0 |
GO:0045410 |
positive regulation of interleukin-12 biosynthetic process(GO:0045084) positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 0.0 |
0.3 |
GO:0000301 |
retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 |
0.1 |
GO:0006957 |
complement activation, alternative pathway(GO:0006957) |
| 0.0 |
0.1 |
GO:0009992 |
cellular water homeostasis(GO:0009992) |
| 0.0 |
0.1 |
GO:0090209 |
negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.0 |
0.1 |
GO:1902259 |
regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.0 |
0.1 |
GO:0070842 |
aggresome assembly(GO:0070842) |
| 0.0 |
0.1 |
GO:0070633 |
transepithelial transport(GO:0070633) |
| 0.0 |
0.0 |
GO:0032875 |
regulation of DNA endoreduplication(GO:0032875) DNA endoreduplication(GO:0042023) |
| 0.0 |
0.1 |
GO:2000009 |
negative regulation of protein localization to cell surface(GO:2000009) |
| 0.0 |
0.1 |
GO:0061368 |
behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 |
0.0 |
GO:0018171 |
peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 |
0.1 |
GO:0042711 |
maternal behavior(GO:0042711) |
| 0.0 |
0.0 |
GO:2000049 |
positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 |
0.1 |
GO:0031536 |
positive regulation of exit from mitosis(GO:0031536) |
| 0.0 |
0.8 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 |
0.1 |
GO:0033292 |
T-tubule organization(GO:0033292) |
| 0.0 |
0.2 |
GO:0003334 |
keratinocyte development(GO:0003334) |
| 0.0 |
0.0 |
GO:0034310 |
primary alcohol catabolic process(GO:0034310) |
| 0.0 |
0.0 |
GO:0008626 |
granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 |
0.1 |
GO:0060371 |
response to pyrethroid(GO:0046684) regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) |
| 0.0 |
0.4 |
GO:0061512 |
protein localization to cilium(GO:0061512) |
| 0.0 |
0.1 |
GO:0007296 |
vitellogenesis(GO:0007296) |
| 0.0 |
0.1 |
GO:0000188 |
inactivation of MAPK activity(GO:0000188) |
| 0.0 |
0.1 |
GO:0071625 |
vocalization behavior(GO:0071625) |
| 0.0 |
0.0 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 |
0.0 |
GO:0072385 |
minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 |
0.0 |
GO:0021852 |
pyramidal neuron migration(GO:0021852) |
| 0.0 |
0.1 |
GO:0089711 |
L-glutamate transmembrane transport(GO:0089711) |
| 0.0 |
0.1 |
GO:0046856 |
phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 |
0.0 |
GO:0042940 |
D-amino acid transport(GO:0042940) |
| 0.0 |
0.0 |
GO:0060763 |
mammary duct terminal end bud growth(GO:0060763) |
| 0.0 |
0.2 |
GO:0086010 |
membrane depolarization during action potential(GO:0086010) |
| 0.0 |
0.1 |
GO:0050858 |
negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) |
| 0.0 |
0.0 |
GO:0044539 |
long-chain fatty acid import(GO:0044539) |
| 0.0 |
0.2 |
GO:0043248 |
proteasome assembly(GO:0043248) |
| 0.0 |
0.1 |
GO:0060044 |
negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.0 |
0.0 |
GO:2000642 |
negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.0 |
0.0 |
GO:0048102 |
autophagic cell death(GO:0048102) |
| 0.0 |
0.1 |
GO:0014807 |
regulation of somitogenesis(GO:0014807) |
| 0.0 |
0.5 |
GO:0002224 |
toll-like receptor signaling pathway(GO:0002224) |
| 0.0 |
0.1 |
GO:0048172 |
regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 |
0.1 |
GO:0051101 |
regulation of DNA binding(GO:0051101) |
| 0.0 |
0.1 |
GO:0014742 |
positive regulation of cardiac muscle hypertrophy(GO:0010613) positive regulation of muscle hypertrophy(GO:0014742) |
| 0.0 |
0.1 |
GO:0060178 |
regulation of exocyst localization(GO:0060178) |
| 0.0 |
0.2 |
GO:0034063 |
stress granule assembly(GO:0034063) |
| 0.0 |
0.2 |
GO:1902187 |
negative regulation of viral release from host cell(GO:1902187) |