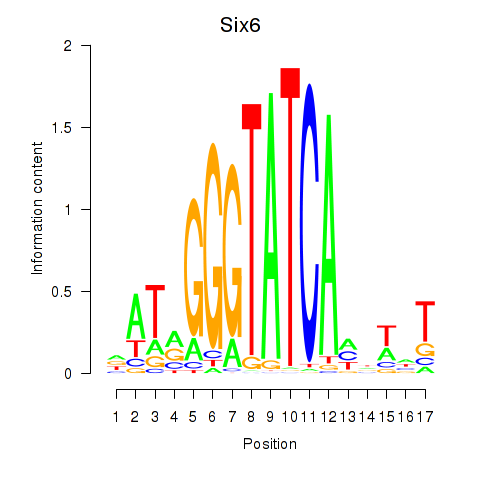
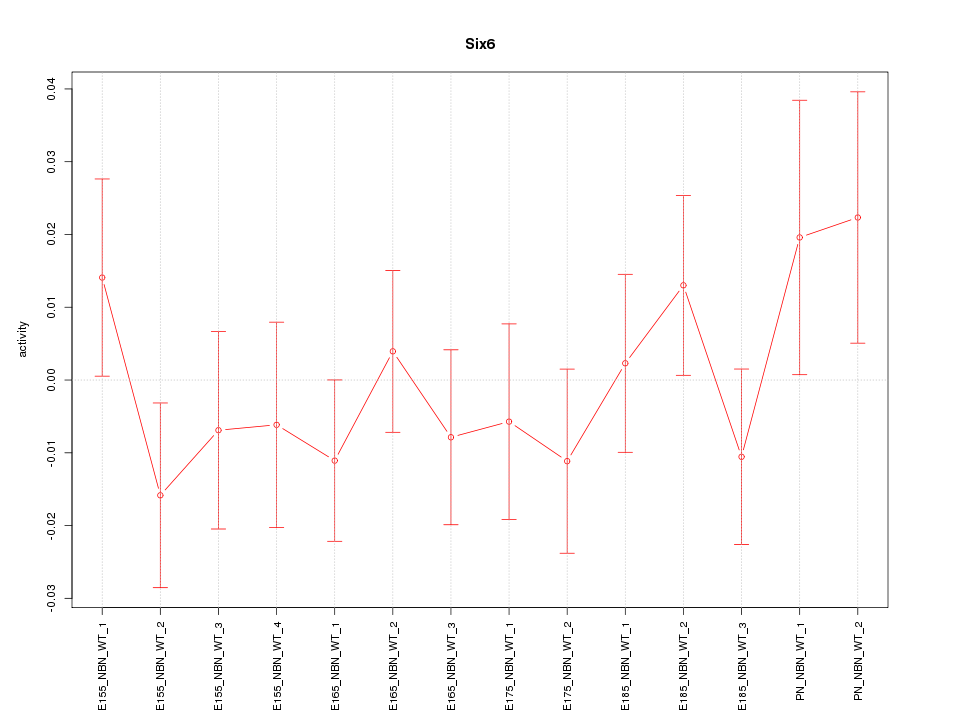
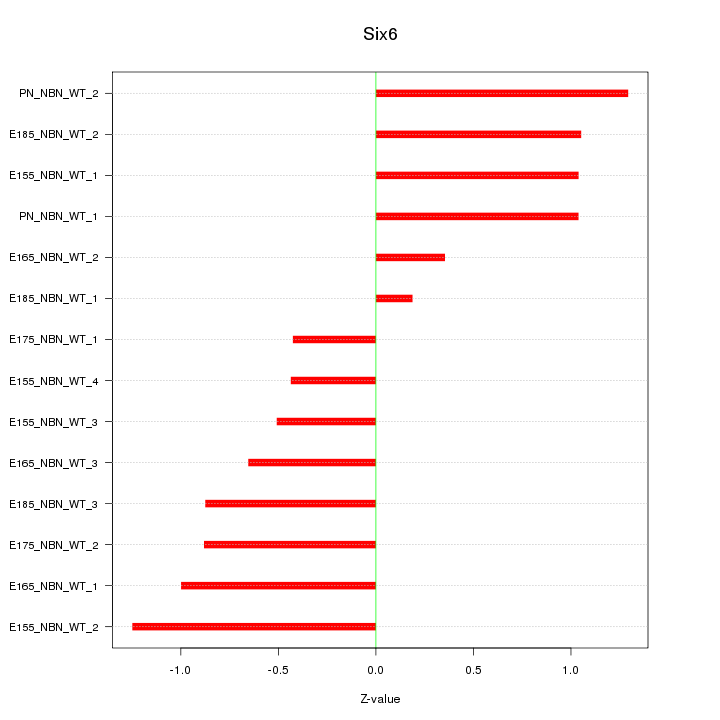
| 0.8 |
3.8 |
GO:0034441 |
plasma lipoprotein particle oxidation(GO:0034441) |
| 0.4 |
1.5 |
GO:0070318 |
positive regulation of G0 to G1 transition(GO:0070318) |
| 0.3 |
2.8 |
GO:0051923 |
sulfation(GO:0051923) |
| 0.2 |
1.0 |
GO:0097393 |
post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.2 |
1.2 |
GO:0007386 |
compartment pattern specification(GO:0007386) |
| 0.2 |
1.0 |
GO:0007089 |
traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.2 |
0.6 |
GO:2000338 |
positive regulation of interleukin-23 production(GO:0032747) chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.2 |
0.7 |
GO:0048539 |
bone marrow development(GO:0048539) |
| 0.2 |
0.8 |
GO:0070236 |
negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.2 |
0.7 |
GO:0006776 |
vitamin A metabolic process(GO:0006776) |
| 0.2 |
0.8 |
GO:0051661 |
maintenance of centrosome location(GO:0051661) |
| 0.1 |
0.7 |
GO:0060414 |
aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 |
1.0 |
GO:0071474 |
cellular hyperosmotic response(GO:0071474) |
| 0.1 |
1.3 |
GO:0030388 |
fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 |
0.4 |
GO:0048496 |
maintenance of organ identity(GO:0048496) |
| 0.1 |
0.7 |
GO:2000002 |
negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.1 |
0.3 |
GO:0002314 |
germinal center B cell differentiation(GO:0002314) |
| 0.1 |
0.8 |
GO:0042760 |
very long-chain fatty acid catabolic process(GO:0042760) |
| 0.1 |
0.8 |
GO:0000712 |
resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 |
0.3 |
GO:0097428 |
protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 |
0.3 |
GO:0060032 |
notochord regression(GO:0060032) |
| 0.1 |
0.5 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 |
0.3 |
GO:0072488 |
ammonium transmembrane transport(GO:0072488) |
| 0.1 |
0.8 |
GO:1901673 |
regulation of mitotic spindle assembly(GO:1901673) |
| 0.1 |
0.5 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.1 |
0.4 |
GO:0071294 |
cellular response to zinc ion(GO:0071294) |
| 0.1 |
1.1 |
GO:0003351 |
epithelial cilium movement(GO:0003351) |
| 0.1 |
0.2 |
GO:0044849 |
estrous cycle(GO:0044849) |
| 0.0 |
0.4 |
GO:0097460 |
positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) ferrous iron import into cell(GO:0097460) |
| 0.0 |
0.2 |
GO:0046208 |
spermine catabolic process(GO:0046208) |
| 0.0 |
0.3 |
GO:1903352 |
ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.0 |
0.4 |
GO:2001214 |
positive regulation of vasculogenesis(GO:2001214) |
| 0.0 |
0.7 |
GO:0045954 |
positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.0 |
0.2 |
GO:0030647 |
polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 |
0.9 |
GO:0034315 |
regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 |
0.2 |
GO:0043461 |
proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 |
0.1 |
GO:0031581 |
hemidesmosome assembly(GO:0031581) |
| 0.0 |
0.4 |
GO:0006268 |
DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 |
0.7 |
GO:0006828 |
manganese ion transport(GO:0006828) |
| 0.0 |
0.1 |
GO:0060392 |
negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 |
0.3 |
GO:0006729 |
tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 |
0.3 |
GO:0050957 |
equilibrioception(GO:0050957) |
| 0.0 |
0.3 |
GO:0016127 |
cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 |
0.5 |
GO:0045792 |
negative regulation of cell size(GO:0045792) |
| 0.0 |
0.2 |
GO:0010748 |
negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 |
0.1 |
GO:0031022 |
nuclear migration along microfilament(GO:0031022) |
| 0.0 |
0.5 |
GO:0033194 |
response to hydroperoxide(GO:0033194) |
| 0.0 |
0.1 |
GO:0019614 |
catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.0 |
0.5 |
GO:0006958 |
complement activation, classical pathway(GO:0006958) |
| 0.0 |
2.5 |
GO:0009566 |
fertilization(GO:0009566) |
| 0.0 |
0.1 |
GO:0001580 |
detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 |
0.2 |
GO:0045974 |
miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 |
0.2 |
GO:0030150 |
protein import into mitochondrial matrix(GO:0030150) |
| 0.0 |
0.3 |
GO:0071624 |
positive regulation of granulocyte chemotaxis(GO:0071624) positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 |
0.2 |
GO:0043928 |
exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 |
0.1 |
GO:0032790 |
ribosome disassembly(GO:0032790) |
| 0.0 |
0.6 |
GO:0007026 |
negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 |
0.4 |
GO:0001706 |
endoderm formation(GO:0001706) |
| 0.0 |
0.2 |
GO:0009048 |
dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 |
0.3 |
GO:0048266 |
behavioral response to pain(GO:0048266) |
| 0.0 |
0.1 |
GO:0071372 |
cellular response to follicle-stimulating hormone stimulus(GO:0071372) |