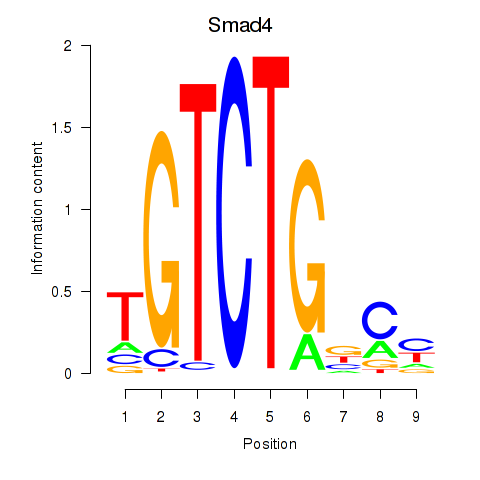
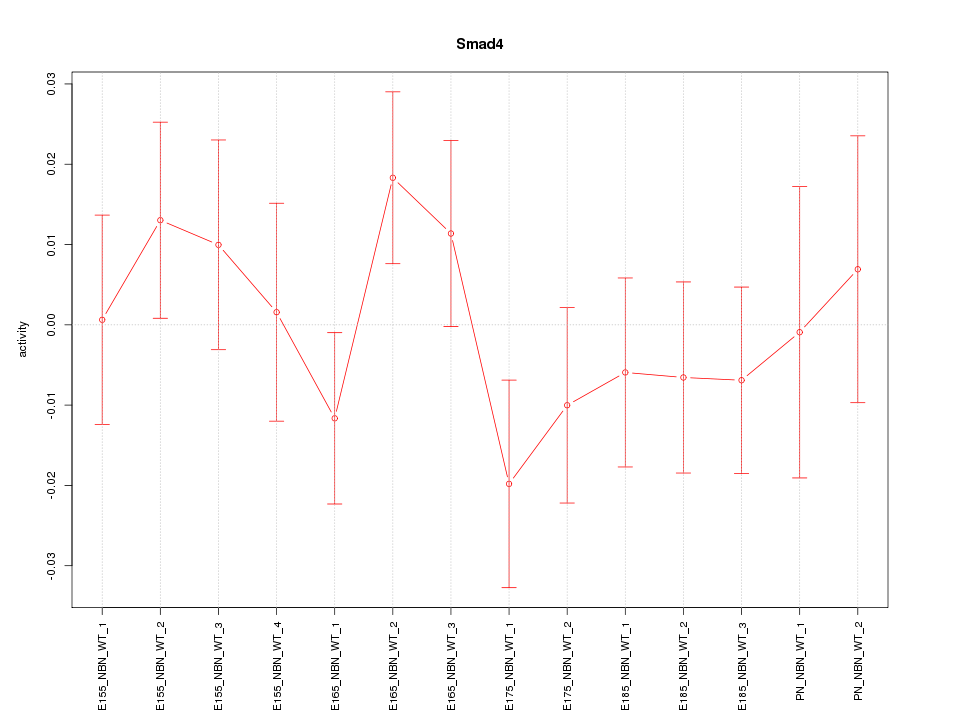
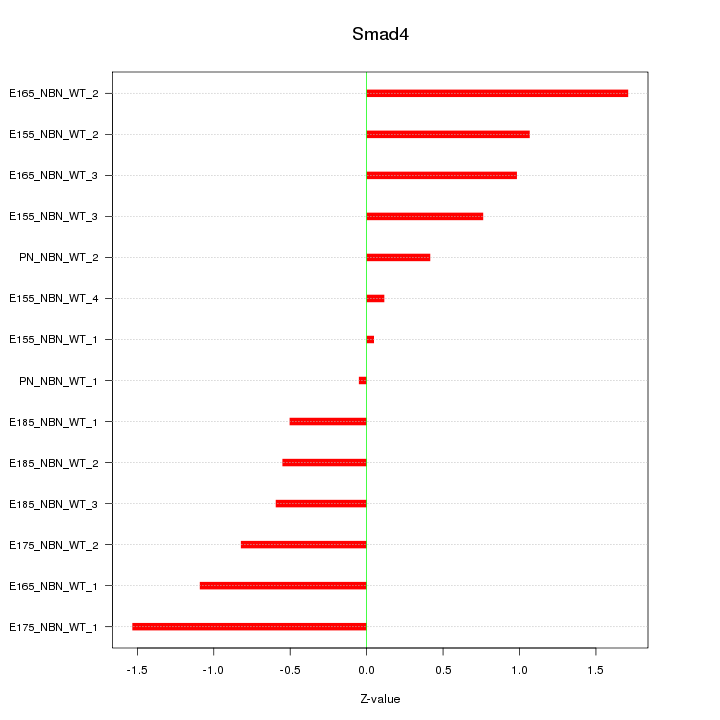
| 0.7 |
3.0 |
GO:2000546 |
positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.4 |
1.3 |
GO:1905065 |
hematopoietic stem cell migration(GO:0035701) positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 0.3 |
1.0 |
GO:1902460 |
regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.2 |
3.1 |
GO:0060159 |
regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.2 |
0.7 |
GO:0003219 |
cardiac right ventricle formation(GO:0003219) |
| 0.2 |
0.8 |
GO:0097709 |
connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.2 |
0.9 |
GO:0014719 |
skeletal muscle satellite cell activation(GO:0014719) |
| 0.2 |
0.3 |
GO:0070885 |
negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.1 |
0.7 |
GO:0021764 |
amygdala development(GO:0021764) |
| 0.1 |
0.4 |
GO:0014005 |
microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.1 |
0.7 |
GO:0060838 |
lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.1 |
0.5 |
GO:1901204 |
regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 0.1 |
0.2 |
GO:0033159 |
negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.1 |
0.3 |
GO:0015889 |
cobalamin transport(GO:0015889) |
| 0.1 |
0.7 |
GO:0019370 |
leukotriene biosynthetic process(GO:0019370) |
| 0.1 |
1.8 |
GO:0048368 |
lateral mesoderm development(GO:0048368) |
| 0.1 |
0.5 |
GO:0010991 |
negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 |
0.4 |
GO:0035331 |
negative regulation of hippo signaling(GO:0035331) |
| 0.1 |
0.2 |
GO:0060414 |
aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 |
0.7 |
GO:0048251 |
elastic fiber assembly(GO:0048251) |
| 0.1 |
0.2 |
GO:1903225 |
negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 |
0.4 |
GO:2000623 |
regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 |
0.1 |
GO:1903351 |
response to dopamine(GO:1903350) cellular response to dopamine(GO:1903351) |
| 0.1 |
0.5 |
GO:0006003 |
fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 |
0.3 |
GO:0003223 |
ventricular compact myocardium morphogenesis(GO:0003223) proprioception(GO:0019230) |
| 0.1 |
0.7 |
GO:0035090 |
maintenance of apical/basal cell polarity(GO:0035090) |
| 0.1 |
0.2 |
GO:0002339 |
B cell selection(GO:0002339) |
| 0.1 |
0.5 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 |
0.3 |
GO:1903300 |
negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 |
0.1 |
GO:2000544 |
cell chemotaxis to fibroblast growth factor(GO:0035766) endothelial cell chemotaxis to fibroblast growth factor(GO:0035768) regulation of cell chemotaxis to fibroblast growth factor(GO:1904847) regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000544) |
| 0.0 |
0.1 |
GO:0032241 |
positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.0 |
0.4 |
GO:0045607 |
regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 |
0.2 |
GO:1903026 |
negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 |
0.5 |
GO:0033690 |
positive regulation of osteoblast proliferation(GO:0033690) |
| 0.0 |
0.2 |
GO:0055071 |
cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.0 |
0.2 |
GO:0070966 |
nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 |
0.2 |
GO:0006172 |
ADP biosynthetic process(GO:0006172) |
| 0.0 |
0.2 |
GO:0006447 |
regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 0.0 |
0.3 |
GO:0033631 |
cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 |
0.7 |
GO:0032967 |
positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 |
0.5 |
GO:0050966 |
detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.0 |
0.3 |
GO:0002227 |
innate immune response in mucosa(GO:0002227) |
| 0.0 |
0.5 |
GO:0070498 |
interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 |
0.1 |
GO:0009298 |
GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 |
1.0 |
GO:0043551 |
regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 |
0.2 |
GO:0036072 |
intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 |
0.5 |
GO:0031295 |
lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 |
0.2 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 |
0.2 |
GO:0071225 |
cellular response to muramyl dipeptide(GO:0071225) |
| 0.0 |
1.2 |
GO:0045600 |
positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 |
0.4 |
GO:0010225 |
response to UV-C(GO:0010225) |
| 0.0 |
0.3 |
GO:0060481 |
lobar bronchus epithelium development(GO:0060481) |
| 0.0 |
0.1 |
GO:0002934 |
desmosome organization(GO:0002934) |
| 0.0 |
0.4 |
GO:0000413 |
protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 |
0.3 |
GO:0000731 |
DNA synthesis involved in DNA repair(GO:0000731) |
| 0.0 |
0.1 |
GO:0071596 |
ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 |
0.0 |
GO:0034476 |
U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 |
0.2 |
GO:0038092 |
nodal signaling pathway(GO:0038092) |
| 0.0 |
0.1 |
GO:0060325 |
face morphogenesis(GO:0060325) |
| 0.0 |
0.2 |
GO:0050434 |
positive regulation of viral transcription(GO:0050434) |
| 0.0 |
0.0 |
GO:0000389 |
mRNA 3'-splice site recognition(GO:0000389) |