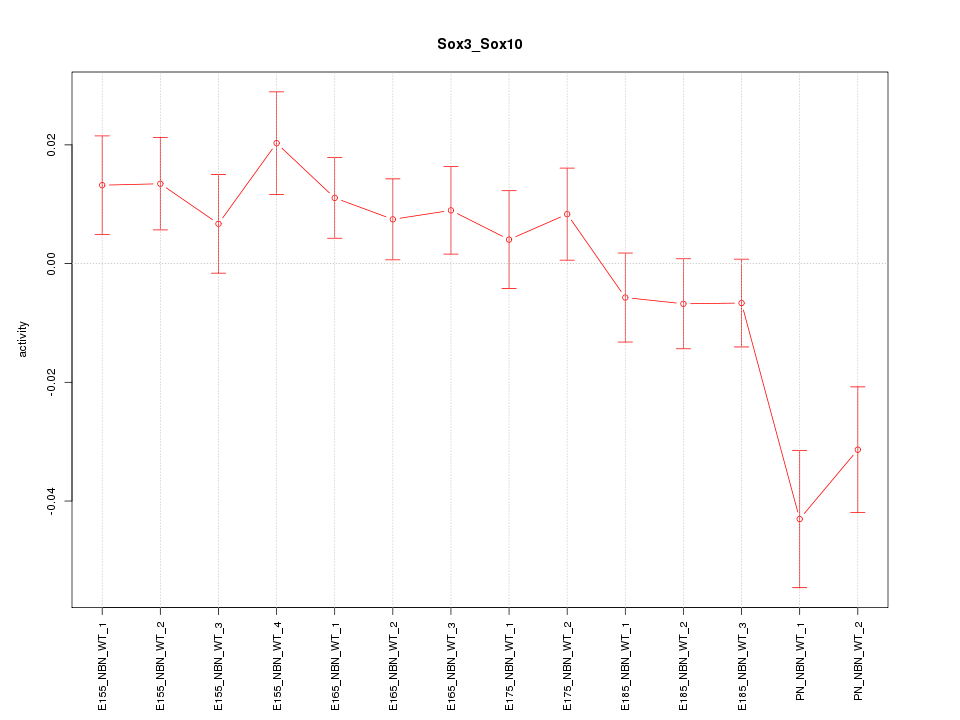
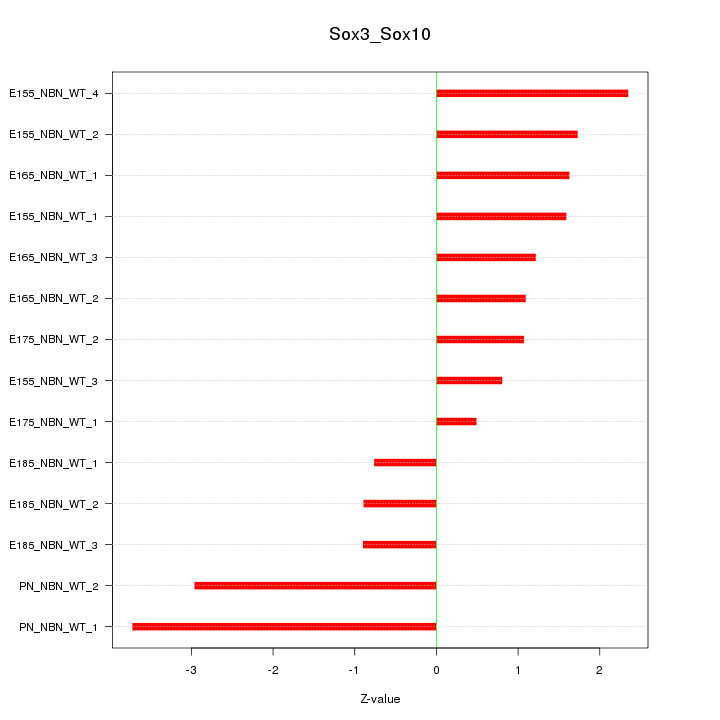
Motif ID: Sox3_Sox10
Z-value: 1.757
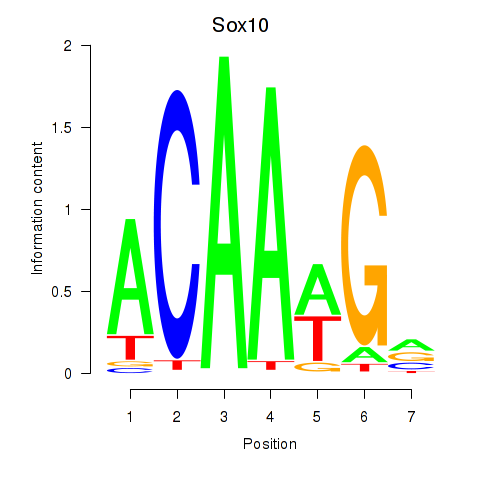
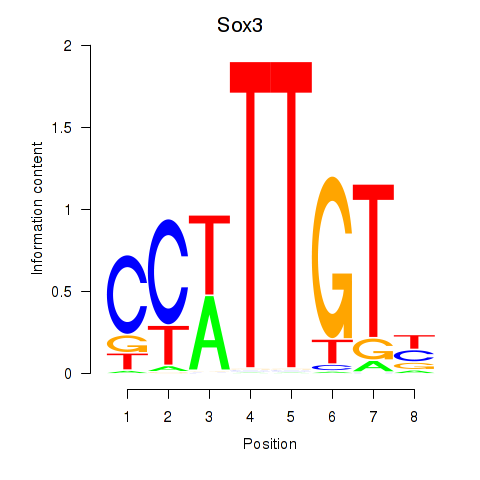
Transcription factors associated with Sox3_Sox10:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Sox10 | ENSMUSG00000033006.9 | Sox10 |
| Sox3 | ENSMUSG00000045179.8 | Sox3 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox10 | mm10_v2_chr15_-_79164477_79164496 | -0.75 | 1.9e-03 | Click! |
| Sox3 | mm10_v2_chrX_-_60893430_60893440 | 0.45 | 1.1e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.7 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 1.4 | 5.6 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 1.1 | 8.8 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 1.0 | 13.6 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 1.0 | 3.1 | GO:0060489 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 1.0 | 6.8 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 1.0 | 2.9 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 1.0 | 2.9 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.9 | 2.7 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.9 | 2.7 | GO:1905065 | hematopoietic stem cell migration(GO:0035701) positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 0.8 | 2.5 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.8 | 3.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.7 | 3.0 | GO:0032289 | central nervous system myelin formation(GO:0032289) cardiac cell fate specification(GO:0060912) |
| 0.7 | 5.6 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.7 | 4.9 | GO:0034047 | regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.7 | 3.3 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.6 | 1.9 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.6 | 5.7 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.6 | 7.0 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.6 | 4.2 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.6 | 2.3 | GO:1904529 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.6 | 1.7 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.6 | 0.6 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.5 | 1.6 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.5 | 2.7 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.5 | 1.6 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.5 | 1.1 | GO:0021814 | cell motility involved in cerebral cortex radial glia guided migration(GO:0021814) |
| 0.5 | 1.6 | GO:0003096 | renal sodium ion transport(GO:0003096) |
| 0.5 | 1.5 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.5 | 9.1 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.5 | 2.0 | GO:2000705 | regulation of dense core granule biogenesis(GO:2000705) |
| 0.5 | 1.5 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.5 | 1.9 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.5 | 1.9 | GO:2001016 | positive regulation of skeletal muscle cell differentiation(GO:2001016) |
| 0.5 | 2.3 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.4 | 2.2 | GO:0071205 | clustering of voltage-gated potassium channels(GO:0045163) protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.4 | 2.5 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.4 | 2.5 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.4 | 1.7 | GO:0035795 | negative regulation of mitochondrial membrane permeability(GO:0035795) |
| 0.4 | 1.7 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.4 | 3.3 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.4 | 1.2 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.4 | 6.0 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.4 | 2.0 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.4 | 7.8 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.4 | 2.3 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.4 | 5.6 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.4 | 1.1 | GO:0032512 | regulation of protein phosphatase type 2B activity(GO:0032512) |
| 0.4 | 1.1 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.3 | 1.7 | GO:0001661 | conditioned taste aversion(GO:0001661) amygdala development(GO:0021764) |
| 0.3 | 2.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.3 | 1.4 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.3 | 1.3 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.3 | 0.7 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.3 | 1.0 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.3 | 1.3 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.3 | 1.6 | GO:0003105 | negative regulation of glomerular filtration(GO:0003105) |
| 0.3 | 4.8 | GO:0010715 | regulation of extracellular matrix disassembly(GO:0010715) |
| 0.3 | 1.6 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.3 | 1.2 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.3 | 1.8 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.3 | 0.9 | GO:1904049 | regulation of spontaneous neurotransmitter secretion(GO:1904048) negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.3 | 1.5 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.3 | 0.9 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.3 | 2.6 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.3 | 0.9 | GO:0050883 | musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.3 | 3.5 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.3 | 1.4 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.3 | 0.3 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.3 | 3.6 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.3 | 0.8 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.3 | 5.7 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.3 | 0.3 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.2 | 1.0 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.2 | 1.2 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.2 | 1.0 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.2 | 0.7 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.2 | 1.0 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.2 | 0.7 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.2 | 0.5 | GO:0071865 | regulation of apoptotic process in bone marrow(GO:0071865) negative regulation of apoptotic process in bone marrow(GO:0071866) |
| 0.2 | 0.7 | GO:0070428 | regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.2 | 4.9 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.2 | 0.7 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.2 | 0.2 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.2 | 0.6 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.2 | 0.6 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.2 | 0.8 | GO:0006867 | asparagine transport(GO:0006867) glutamine transport(GO:0006868) |
| 0.2 | 0.6 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.2 | 1.5 | GO:0086018 | SA node cell action potential(GO:0086015) SA node cell to atrial cardiac muscle cell signalling(GO:0086018) SA node cell to atrial cardiac muscle cell communication(GO:0086070) |
| 0.2 | 2.8 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.2 | 0.6 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.2 | 0.4 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.2 | 1.2 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.2 | 0.4 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.2 | 0.2 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.2 | 0.8 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.2 | 0.8 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.2 | 1.0 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.2 | 0.4 | GO:1903898 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.2 | 0.7 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.2 | 0.5 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.2 | 3.9 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.2 | 1.6 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.2 | 0.4 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.2 | 0.9 | GO:1904690 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.2 | 0.5 | GO:0032650 | regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) interleukin-1 alpha secretion(GO:0050703) |
| 0.2 | 0.7 | GO:0006547 | histidine metabolic process(GO:0006547) |
| 0.2 | 5.5 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.2 | 0.7 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.2 | 1.0 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.2 | 0.9 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.2 | 0.5 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.2 | 0.7 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900175) positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) positive regulation of mesoderm development(GO:2000382) |
| 0.2 | 1.7 | GO:0009186 | deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.2 | 0.5 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.2 | 1.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.2 | 1.4 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.2 | 0.8 | GO:0001865 | NK T cell differentiation(GO:0001865) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.2 | 0.6 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.2 | 0.9 | GO:1903753 | negative regulation of p38MAPK cascade(GO:1903753) |
| 0.2 | 0.9 | GO:0018377 | N-terminal protein myristoylation(GO:0006499) protein myristoylation(GO:0018377) |
| 0.2 | 2.7 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.2 | 0.8 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.2 | 0.8 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.2 | 0.5 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.1 | 0.4 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.1 | 0.7 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 | 0.6 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.1 | 0.3 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.1 | 2.3 | GO:1901629 | regulation of presynaptic membrane organization(GO:1901629) |
| 0.1 | 0.6 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 11.5 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.1 | 1.1 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 2.5 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 0.6 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 1.0 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 0.8 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 0.9 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 1.0 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.1 | 0.8 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 0.4 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 0.8 | GO:0009133 | nucleoside diphosphate biosynthetic process(GO:0009133) ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.1 | 1.6 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 0.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.7 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 0.2 | GO:0060025 | regulation of synaptic activity(GO:0060025) |
| 0.1 | 1.0 | GO:0010878 | cholesterol storage(GO:0010878) steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.1 | 0.5 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.1 | 1.2 | GO:0071225 | cellular response to muramyl dipeptide(GO:0071225) |
| 0.1 | 0.8 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.1 | 0.3 | GO:0035984 | response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.1 | 2.2 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.1 | 0.8 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.3 | GO:0048352 | neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) |
| 0.1 | 0.5 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.6 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.1 | 0.5 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.1 | 5.1 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.1 | 1.9 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 0.4 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 0.6 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.1 | 0.3 | GO:1902950 | regulation of dendritic spine maintenance(GO:1902950) positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.1 | 0.5 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 0.4 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.1 | 1.3 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 0.5 | GO:0031652 | positive regulation of heat generation(GO:0031652) |
| 0.1 | 0.4 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.5 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.1 | 0.5 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.1 | 0.3 | GO:0003149 | membranous septum morphogenesis(GO:0003149) |
| 0.1 | 1.3 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 0.7 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 2.1 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 0.6 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.1 | 0.3 | GO:0015838 | amino-acid betaine transport(GO:0015838) |
| 0.1 | 0.3 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.1 | 0.4 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 0.6 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.1 | 1.1 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.1 | 0.5 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.1 | 0.3 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.1 | 0.6 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.1 | 0.4 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.1 | 0.5 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.1 | 0.9 | GO:1900112 | regulation of histone H3-K9 trimethylation(GO:1900112) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 | 0.4 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.1 | 0.7 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.1 | 0.4 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.1 | 0.3 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 1.2 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.1 | 0.3 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.1 | 0.8 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.1 | 0.3 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.1 | 2.3 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.1 | 0.4 | GO:0050912 | detection of chemical stimulus involved in sensory perception(GO:0050907) detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.1 | 0.8 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 | 0.7 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 0.2 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.1 | 0.2 | GO:1905154 | negative regulation of tumor necrosis factor secretion(GO:1904468) negative regulation of membrane invagination(GO:1905154) |
| 0.1 | 0.3 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.2 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.1 | 0.8 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.1 | 3.4 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 0.6 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 0.8 | GO:0060013 | righting reflex(GO:0060013) |
| 0.1 | 2.1 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 0.4 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.1 | 0.5 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.1 | 0.4 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 0.4 | GO:0015692 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.1 | 0.8 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 0.3 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 | 1.3 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 8.4 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.1 | 0.6 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 1.4 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.1 | 0.2 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.1 | 2.3 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.1 | 1.5 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.1 | 0.7 | GO:0006337 | nucleosome disassembly(GO:0006337) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 0.7 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.1 | 0.3 | GO:1904754 | positive regulation of vascular associated smooth muscle cell migration(GO:1904754) |
| 0.1 | 4.1 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 0.1 | GO:1903351 | response to dopamine(GO:1903350) cellular response to dopamine(GO:1903351) regulation of sodium:potassium-exchanging ATPase activity(GO:1903406) |
| 0.1 | 0.5 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.1 | 2.6 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 0.3 | GO:0061045 | negative regulation of wound healing(GO:0061045) |
| 0.1 | 0.5 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 0.6 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.1 | 1.1 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 0.1 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.1 | 0.3 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 0.6 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.1 | 0.5 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 0.2 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 | 0.2 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.1 | 2.0 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.1 | 2.0 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.1 | 0.5 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.1 | 0.2 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.1 | 0.5 | GO:0002437 | inflammatory response to antigenic stimulus(GO:0002437) |
| 0.1 | 0.3 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.1 | 0.9 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.1 | 1.6 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 1.7 | GO:0090004 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 0.7 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.7 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 0.9 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.1 | 0.8 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.5 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.1 | 0.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.1 | 0.2 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 0.3 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.0 | 1.0 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.3 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 1.4 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.0 | 1.1 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.3 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.0 | 0.4 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.6 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.6 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.3 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.5 | GO:0021540 | corpus callosum morphogenesis(GO:0021540) |
| 0.0 | 0.6 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.7 | GO:0048305 | immunoglobulin secretion(GO:0048305) |
| 0.0 | 0.2 | GO:0031055 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.3 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.0 | 2.7 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.3 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 1.2 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.0 | 0.5 | GO:0023019 | signal transduction involved in regulation of gene expression(GO:0023019) |
| 0.0 | 1.0 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.2 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 | 1.0 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 2.8 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.6 | GO:0044783 | mitotic G1 DNA damage checkpoint(GO:0031571) G1 DNA damage checkpoint(GO:0044783) |
| 0.0 | 0.2 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.3 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.6 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 1.2 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.3 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.6 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.6 | GO:1903427 | negative regulation of reactive oxygen species biosynthetic process(GO:1903427) |
| 0.0 | 0.4 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 2.0 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.2 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.4 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.5 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.4 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 1.3 | GO:1900046 | regulation of blood coagulation(GO:0030193) regulation of hemostasis(GO:1900046) |
| 0.0 | 0.7 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 3.7 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.6 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.3 | GO:0086003 | cardiac muscle cell contraction(GO:0086003) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0021649 | vestibulocochlear nerve morphogenesis(GO:0021648) vestibulocochlear nerve structural organization(GO:0021649) cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) ganglion morphogenesis(GO:0061552) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) neural crest cell migration involved in autonomic nervous system development(GO:1901166) facioacoustic ganglion development(GO:1903375) dorsal root ganglion morphogenesis(GO:1904835) dorsal root ganglion development(GO:1990791) |
| 0.0 | 0.7 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.0 | 0.5 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 | 1.3 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.1 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.0 | 0.3 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.1 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 | 0.5 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.0 | 0.6 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.3 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.1 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.1 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 | 0.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.1 | GO:0071680 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 0.3 | GO:0034377 | plasma lipoprotein particle assembly(GO:0034377) |
| 0.0 | 0.6 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.1 | GO:0045213 | neurotransmitter receptor metabolic process(GO:0045213) |
| 0.0 | 0.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.8 | GO:0030148 | sphingolipid biosynthetic process(GO:0030148) |
| 0.0 | 1.2 | GO:0030517 | negative regulation of axon extension(GO:0030517) |
| 0.0 | 0.5 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.0 | 0.2 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.5 | GO:0035904 | aorta development(GO:0035904) |
| 0.0 | 0.6 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.3 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 3.4 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 0.5 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.1 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 1.5 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 | 0.3 | GO:0000394 | RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.3 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.2 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.7 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.1 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.0 | 0.3 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.0 | 1.4 | GO:0010950 | positive regulation of endopeptidase activity(GO:0010950) |
| 0.0 | 0.1 | GO:0000022 | mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 1.2 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 0.1 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.0 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.1 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.2 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.5 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.0 | 0.1 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.8 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.2 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.2 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.4 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.2 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.0 | 0.1 | GO:0046036 | GTP biosynthetic process(GO:0006183) CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.0 | 0.2 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.0 | 0.3 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.0 | 0.1 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.1 | GO:0001773 | myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 1.2 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.0 | 0.6 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.1 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 1.0 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.4 | GO:0007059 | chromosome segregation(GO:0007059) |
| 0.0 | 0.2 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.0 | 0.0 | GO:0021933 | radial glia guided migration of cerebellar granule cell(GO:0021933) |
| 0.0 | 0.3 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.5 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.5 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.3 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.1 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 1.5 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 1.0 | GO:0051028 | mRNA transport(GO:0051028) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 1.2 | 11.0 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.8 | 4.8 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.8 | 3.9 | GO:0008623 | CHRAC(GO:0008623) |
| 0.8 | 3.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.6 | 1.9 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.5 | 11.1 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.5 | 1.9 | GO:0031673 | H zone(GO:0031673) |
| 0.4 | 10.5 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.4 | 1.7 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.4 | 2.5 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.4 | 1.7 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.4 | 1.2 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) |
| 0.4 | 3.8 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.4 | 7.7 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.3 | 1.4 | GO:0043259 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) |
| 0.3 | 7.0 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.3 | 10.9 | GO:0043034 | costamere(GO:0043034) |
| 0.3 | 2.5 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.3 | 4.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 1.2 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.2 | 2.6 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.2 | 3.0 | GO:0016342 | catenin complex(GO:0016342) |
| 0.2 | 0.5 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.2 | 3.6 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.2 | 1.1 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.2 | 1.7 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.2 | 1.5 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.2 | 1.9 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.2 | 0.4 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.2 | 0.9 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.2 | 1.2 | GO:0071437 | invadopodium(GO:0071437) |
| 0.2 | 1.9 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.2 | 0.7 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.2 | 0.7 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.2 | 1.3 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.2 | 1.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 2.5 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.4 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 0.8 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 2.0 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.1 | 3.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 2.1 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.1 | 0.4 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.1 | 2.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 5.4 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 1.4 | GO:0031105 | septin complex(GO:0031105) |
| 0.1 | 0.6 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 0.3 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.1 | 3.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 2.1 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.1 | 2.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 0.3 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 4.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.8 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 0.5 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 0.5 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 3.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 1.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.3 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.1 | 1.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 1.1 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.1 | 0.3 | GO:0071914 | prominosome(GO:0071914) |
| 0.1 | 0.5 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 1.5 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 1.2 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 16.0 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 1.0 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 1.7 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 4.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 4.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.3 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 4.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 1.0 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.1 | 1.0 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.4 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 3.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 2.1 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 0.7 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 4.2 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 0.2 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.1 | 0.7 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.1 | 5.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 0.2 | GO:0044308 | axonal spine(GO:0044308) |
| 0.1 | 5.4 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 0.3 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 1.3 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 1.1 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.3 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.2 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 1.6 | GO:0015630 | microtubule cytoskeleton(GO:0015630) |
| 0.0 | 0.3 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 2.1 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.8 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.1 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 0.4 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.1 | GO:1990047 | spindle matrix(GO:1990047) |
| 0.0 | 3.0 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.8 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.3 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 1.8 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.3 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.3 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.1 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 1.9 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.2 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.2 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 0.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.8 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 1.8 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 1.4 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.8 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.1 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.3 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.7 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.1 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 1.0 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.5 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.0 | 0.3 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 7.2 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 1.2 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 2.1 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.4 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 1.9 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.6 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.2 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.4 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.4 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.2 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 6.0 | GO:0005829 | cytosol(GO:0005829) |
| 0.0 | 0.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.2 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.9 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.3 | GO:0098797 | plasma membrane protein complex(GO:0098797) |
| 0.0 | 0.4 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.9 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.6 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.5 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.4 | GO:0032040 | small-subunit processome(GO:0032040) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 10.6 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 1.0 | 6.9 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.9 | 6.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.8 | 3.1 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.8 | 2.3 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.6 | 5.7 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.5 | 1.6 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.5 | 1.6 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.5 | 2.6 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.5 | 4.6 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.5 | 2.5 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.4 | 7.3 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.4 | 1.2 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.4 | 3.5 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.4 | 1.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.4 | 9.1 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.4 | 1.5 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.3 | 4.9 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.3 | 1.0 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.3 | 1.7 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.3 | 2.7 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.3 | 16.3 | GO:0004120 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) photoreceptor cyclic-nucleotide phosphodiesterase activity(GO:0004120) 7,8-dihydro-D-neopterin 2',3'-cyclic phosphate phosphodiesterase activity(GO:0044688) inositol phosphosphingolipid phospholipase activity(GO:0052712) inositol phosphorylceramide phospholipase activity(GO:0052713) mannosyl-inositol phosphorylceramide phospholipase activity(GO:0052714) mannosyl-diinositol phosphorylceramide phospholipase activity(GO:0052715) |
| 0.3 | 1.0 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.3 | 2.2 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.3 | 1.9 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.3 | 1.2 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.3 | 1.2 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.3 | 0.9 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.3 | 3.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.3 | 2.7 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.3 | 1.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 0.5 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.2 | 1.2 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.2 | 7.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 0.7 | GO:0015173 | hydrogen:amino acid symporter activity(GO:0005280) aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.2 | 4.3 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.2 | 0.7 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.2 | 8.3 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.2 | 2.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 2.1 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.2 | 0.7 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.2 | 6.9 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.2 | 1.3 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.2 | 0.6 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.2 | 1.7 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.2 | 0.6 | GO:0050827 | toxin receptor binding(GO:0050827) |
| 0.2 | 1.6 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.2 | 5.3 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.2 | 1.7 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.2 | 0.8 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.2 | 1.5 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.2 | 0.8 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.2 | 1.3 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.2 | 4.2 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.2 | 1.3 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.2 | 2.4 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.2 | 6.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 1.0 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.2 | 1.4 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.2 | 0.8 | GO:0015186 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.2 | 6.1 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.2 | 1.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.2 | 4.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 0.6 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 0.7 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.4 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.1 | 0.9 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 0.4 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 0.4 | GO:0004127 | cytidylate kinase activity(GO:0004127) uridylate kinase activity(GO:0009041) |
| 0.1 | 1.4 | GO:0052872 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) phenanthrene 9,10-monooxygenase activity(GO:0018636) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) tocotrienol omega-hydroxylase activity(GO:0052872) thalianol hydroxylase activity(GO:0080014) |
| 0.1 | 0.8 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 1.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 1.9 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.7 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 0.5 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 1.4 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 2.9 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 1.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 3.3 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 1.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 1.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 2.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.1 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.1 | 0.5 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 1.0 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 1.0 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.2 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.1 | 0.7 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 1.9 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 2.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 0.3 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 1.5 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 2.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.8 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.1 | 0.3 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.1 | 0.4 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.4 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.1 | 1.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.9 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.4 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.1 | 1.9 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.1 | 1.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.6 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.4 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.1 | 0.4 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 1.6 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.8 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 2.0 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 0.9 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 1.5 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.1 | 1.9 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.8 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 1.0 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.1 | 1.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.1 | 0.2 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 0.8 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 1.0 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 4.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 0.4 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 0.9 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 11.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 0.2 | GO:0098634 | protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.4 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 1.9 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 0.6 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.3 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.1 | 3.2 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.1 | 0.4 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 2.2 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 0.3 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.1 | 0.4 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 0.5 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.1 | 1.7 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.8 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 0.3 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 1.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 1.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 0.4 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 0.1 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.1 | 1.3 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 0.8 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 1.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 2.3 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.1 | 0.5 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 1.5 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.4 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 1.0 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.5 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 0.5 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.6 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 2.3 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 1.8 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) |
| 0.1 | 0.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 2.5 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 0.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.2 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.1 | 1.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 10.9 | GO:0003779 | actin binding(GO:0003779) |
| 0.1 | 0.4 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
| 0.1 | 0.6 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.1 | 0.2 | GO:0071532 | ornithine decarboxylase inhibitor activity(GO:0008073) ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.9 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.4 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.2 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.4 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0019107 | myristoyltransferase activity(GO:0019107) |
| 0.0 | 0.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.5 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.5 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.5 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 1.0 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 2.2 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.7 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.3 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.3 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.7 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.3 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.1 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 2.7 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.8 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 1.4 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.5 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 3.1 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 8.3 | GO:0042277 | peptide binding(GO:0042277) |
| 0.0 | 0.4 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.3 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.7 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.5 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.6 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 6.6 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 3.2 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 2.0 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.3 | GO:0016653 | oxidoreductase activity, acting on NAD(P)H, heme protein as acceptor(GO:0016653) |
| 0.0 | 0.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 3.9 | GO:0043774 | UDP-N-acetylmuramoylalanyl-D-glutamyl-2,6-diaminopimelate-D-alanyl-D-alanine ligase activity(GO:0008766) ribosomal S6-glutamic acid ligase activity(GO:0018169) coenzyme F420-0 gamma-glutamyl ligase activity(GO:0043773) coenzyme F420-2 alpha-glutamyl ligase activity(GO:0043774) protein-glycine ligase activity(GO:0070735) protein-glycine ligase activity, initiating(GO:0070736) protein-glycine ligase activity, elongating(GO:0070737) tubulin-glycine ligase activity(GO:0070738) |
| 0.0 | 0.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 2.8 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.3 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.5 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 1.4 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.1 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.1 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.0 | 0.3 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.0 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 2.7 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.2 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.2 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:2001070 | glycerophosphocholine phosphodiesterase activity(GO:0047389) starch binding(GO:2001070) |
| 0.0 | 0.6 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 1.7 | GO:0101005 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.5 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.0 | 1.4 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 0.1 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.3 | GO:0043747 | protein-N-terminal asparagine amidohydrolase activity(GO:0008418) UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase activity(GO:0008759) iprodione amidohydrolase activity(GO:0018748) (3,5-dichlorophenylurea)acetate amidohydrolase activity(GO:0018749) 4'-(2-hydroxyisopropyl)phenylurea amidohydrolase activity(GO:0034571) didemethylisoproturon amidohydrolase activity(GO:0034573) N-isopropylacetanilide amidohydrolase activity(GO:0034576) N-cyclohexylformamide amidohydrolase activity(GO:0034781) isonicotinic acid hydrazide hydrolase activity(GO:0034876) cis-aconitamide amidase activity(GO:0034882) gamma-N-formylaminovinylacetate hydrolase activity(GO:0034885) N2-acetyl-L-lysine deacetylase activity(GO:0043747) O-succinylbenzoate synthase activity(GO:0043748) indoleacetamide hydrolase activity(GO:0043864) N-acetylcitrulline deacetylase activity(GO:0043909) N-acetylgalactosamine-6-phosphate deacetylase activity(GO:0047419) diacetylchitobiose deacetylase activity(GO:0052773) chitooligosaccharide deacetylase activity(GO:0052790) |
| 0.0 | 0.2 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.3 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.0 | 0.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 1.5 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.1 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.0 | 0.6 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.0 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.9 | GO:0019210 | kinase inhibitor activity(GO:0019210) |
| 0.0 | 0.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 2.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.3 | GO:0034930 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) heparan sulfate 2-O-sulfotransferase activity(GO:0004394) HNK-1 sulfotransferase activity(GO:0016232) heparan sulfate 6-O-sulfotransferase activity(GO:0017095) trans-9R,10R-dihydrodiolphenanthrene sulfotransferase activity(GO:0018721) 1-phenanthrol sulfotransferase activity(GO:0018722) 3-phenanthrol sulfotransferase activity(GO:0018723) 4-phenanthrol sulfotransferase activity(GO:0018724) trans-3,4-dihydrodiolphenanthrene sulfotransferase activity(GO:0018725) 9-phenanthrol sulfotransferase activity(GO:0018726) 2-phenanthrol sulfotransferase activity(GO:0018727) phenanthrol sulfotransferase activity(GO:0019111) 1-hydroxypyrene sulfotransferase activity(GO:0034930) proteoglycan sulfotransferase activity(GO:0050698) cholesterol sulfotransferase activity(GO:0051922) hydroxyjasmonate sulfotransferase activity(GO:0080131) |