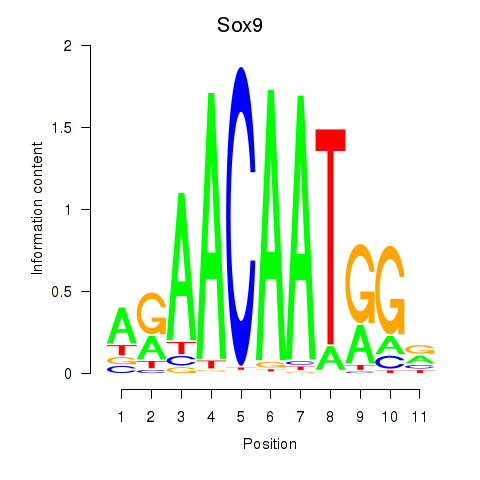
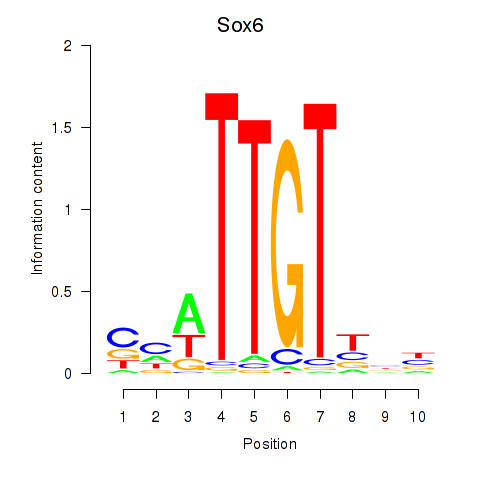
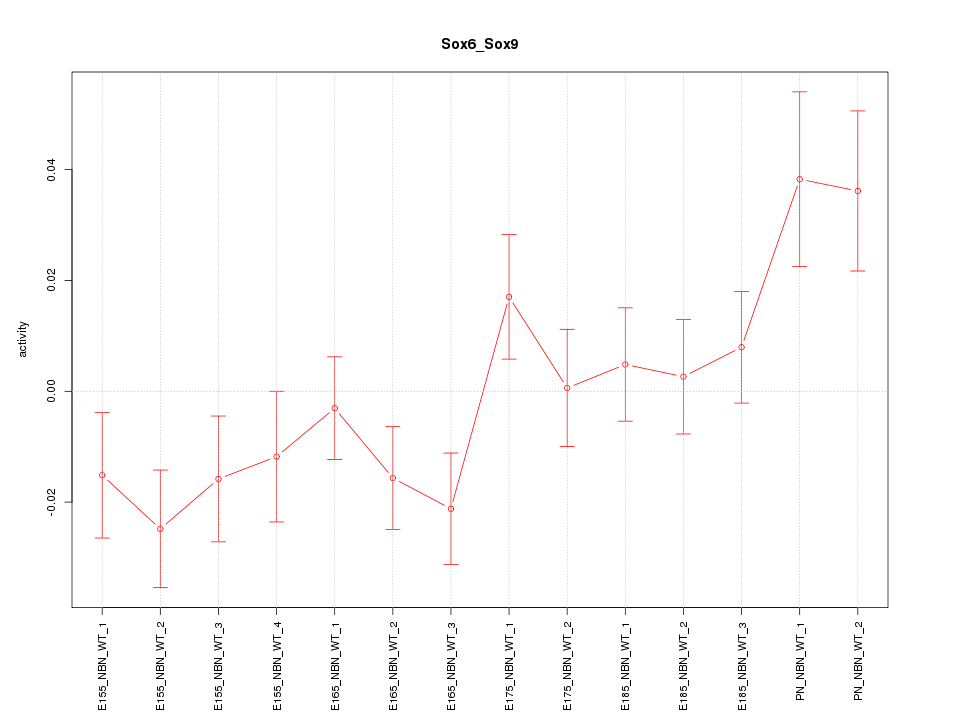
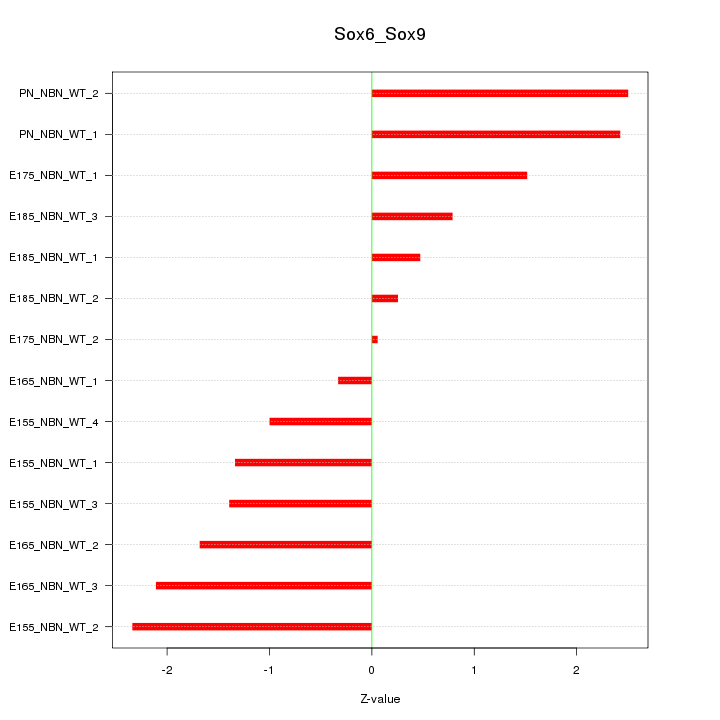
| 1.1 |
3.2 |
GO:0021837 |
motogenic signaling involved in postnatal olfactory bulb interneuron migration(GO:0021837) positive regulation of mitotic cell cycle DNA replication(GO:1903465) |
| 0.9 |
5.6 |
GO:0060665 |
regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.8 |
2.5 |
GO:0042560 |
10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.8 |
3.4 |
GO:0003357 |
noradrenergic neuron differentiation(GO:0003357) |
| 0.8 |
3.9 |
GO:2000298 |
regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.7 |
7.4 |
GO:1903608 |
protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.7 |
2.6 |
GO:0050904 |
diapedesis(GO:0050904) |
| 0.6 |
2.5 |
GO:1904425 |
negative regulation of GTP binding(GO:1904425) |
| 0.5 |
3.7 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.5 |
3.4 |
GO:0071699 |
olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.4 |
2.2 |
GO:0047484 |
regulation of response to osmotic stress(GO:0047484) |
| 0.4 |
1.9 |
GO:0046684 |
response to pyrethroid(GO:0046684) |
| 0.4 |
0.8 |
GO:0035426 |
extracellular matrix-cell signaling(GO:0035426) |
| 0.4 |
3.7 |
GO:0034983 |
peptidyl-lysine deacetylation(GO:0034983) |
| 0.3 |
1.0 |
GO:0090285 |
negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.3 |
2.4 |
GO:0061042 |
vascular wound healing(GO:0061042) |
| 0.3 |
1.0 |
GO:0050929 |
corticospinal neuron axon guidance(GO:0021966) corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.3 |
2.5 |
GO:0033353 |
S-adenosylmethionine cycle(GO:0033353) |
| 0.3 |
0.9 |
GO:2001015 |
G1 to G0 transition involved in cell differentiation(GO:0070315) negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.3 |
2.5 |
GO:0021546 |
rhombomere development(GO:0021546) |
| 0.3 |
1.8 |
GO:0003406 |
retinal pigment epithelium development(GO:0003406) |
| 0.3 |
1.8 |
GO:0098700 |
aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.3 |
1.9 |
GO:1902969 |
mitotic DNA replication(GO:1902969) |
| 0.3 |
2.9 |
GO:0051923 |
sulfation(GO:0051923) |
| 0.3 |
1.8 |
GO:0061304 |
retinal blood vessel morphogenesis(GO:0061304) |
| 0.3 |
1.0 |
GO:1900220 |
semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.2 |
1.0 |
GO:2000048 |
negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.2 |
0.7 |
GO:0048014 |
Tie signaling pathway(GO:0048014) |
| 0.2 |
0.7 |
GO:0043696 |
dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.2 |
0.7 |
GO:0035441 |
cell migration involved in vasculogenesis(GO:0035441) |
| 0.2 |
1.4 |
GO:1900246 |
positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.2 |
1.1 |
GO:0032534 |
regulation of microvillus assembly(GO:0032534) |
| 0.2 |
0.9 |
GO:0045409 |
negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.2 |
2.6 |
GO:0031274 |
positive regulation of pseudopodium assembly(GO:0031274) |
| 0.2 |
0.6 |
GO:1904059 |
regulation of locomotor rhythm(GO:1904059) |
| 0.2 |
0.8 |
GO:2000383 |
regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.2 |
1.0 |
GO:0097116 |
gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.2 |
2.2 |
GO:0045607 |
regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.2 |
1.0 |
GO:0010792 |
DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.2 |
1.3 |
GO:0030382 |
sperm mitochondrion organization(GO:0030382) |
| 0.2 |
0.7 |
GO:0016071 |
mRNA metabolic process(GO:0016071) |
| 0.2 |
1.2 |
GO:0019532 |
oxalate transport(GO:0019532) |
| 0.2 |
0.7 |
GO:1900740 |
regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.2 |
0.9 |
GO:0097368 |
establishment of Sertoli cell barrier(GO:0097368) |
| 0.2 |
1.3 |
GO:0014807 |
regulation of somitogenesis(GO:0014807) |
| 0.2 |
0.8 |
GO:0034382 |
chylomicron remnant clearance(GO:0034382) positive regulation of lipoprotein lipase activity(GO:0051006) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.2 |
1.1 |
GO:0060903 |
positive regulation of meiosis I(GO:0060903) |
| 0.1 |
1.0 |
GO:0030718 |
germ-line stem cell population maintenance(GO:0030718) |
| 0.1 |
0.4 |
GO:0060217 |
hemangioblast cell differentiation(GO:0060217) |
| 0.1 |
0.9 |
GO:0046007 |
negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 |
0.6 |
GO:0051684 |
maintenance of Golgi location(GO:0051684) |
| 0.1 |
0.4 |
GO:0003192 |
mitral valve formation(GO:0003192) |
| 0.1 |
0.6 |
GO:0045578 |
negative regulation of B cell differentiation(GO:0045578) |
| 0.1 |
0.8 |
GO:1904936 |
cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 |
1.7 |
GO:0032620 |
interleukin-17 production(GO:0032620) |
| 0.1 |
2.0 |
GO:0042346 |
positive regulation of NF-kappaB import into nucleus(GO:0042346) |
| 0.1 |
2.2 |
GO:0035855 |
megakaryocyte development(GO:0035855) |
| 0.1 |
1.8 |
GO:0042759 |
long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 |
1.5 |
GO:0038063 |
collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 |
1.0 |
GO:0010587 |
miRNA catabolic process(GO:0010587) |
| 0.1 |
0.3 |
GO:0036091 |
positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 |
0.5 |
GO:1905049 |
negative regulation of metalloendopeptidase activity(GO:1904684) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.1 |
1.0 |
GO:0046085 |
adenosine metabolic process(GO:0046085) |
| 0.1 |
0.5 |
GO:0035262 |
gonad morphogenesis(GO:0035262) |
| 0.1 |
1.0 |
GO:0032876 |
negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 |
2.5 |
GO:0001573 |
ganglioside metabolic process(GO:0001573) |
| 0.1 |
0.3 |
GO:0038203 |
TORC2 signaling(GO:0038203) |
| 0.1 |
0.8 |
GO:0070574 |
cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 |
1.0 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 |
0.2 |
GO:0090327 |
negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.1 |
0.6 |
GO:0048625 |
myoblast fate commitment(GO:0048625) |
| 0.1 |
0.4 |
GO:0010835 |
regulation of protein ADP-ribosylation(GO:0010835) |
| 0.1 |
1.0 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 |
0.2 |
GO:0060399 |
positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.1 |
0.6 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.1 |
1.3 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 |
0.5 |
GO:0001922 |
B-1 B cell homeostasis(GO:0001922) |
| 0.1 |
0.3 |
GO:0071896 |
protein localization to adherens junction(GO:0071896) |
| 0.1 |
0.4 |
GO:0016127 |
cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 |
0.2 |
GO:0006438 |
valyl-tRNA aminoacylation(GO:0006438) |
| 0.1 |
0.9 |
GO:0046688 |
response to copper ion(GO:0046688) |
| 0.1 |
0.2 |
GO:0014835 |
myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.1 |
0.2 |
GO:1901421 |
positive regulation of response to alcohol(GO:1901421) |
| 0.1 |
0.5 |
GO:0061032 |
visceral serous pericardium development(GO:0061032) |
| 0.1 |
1.8 |
GO:0090162 |
establishment of epithelial cell polarity(GO:0090162) |
| 0.1 |
1.4 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 |
1.2 |
GO:2000310 |
regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.1 |
3.0 |
GO:0006940 |
regulation of smooth muscle contraction(GO:0006940) |
| 0.1 |
0.4 |
GO:0090074 |
negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 |
3.3 |
GO:0070527 |
platelet aggregation(GO:0070527) |
| 0.1 |
0.4 |
GO:0034773 |
histone H4-K20 trimethylation(GO:0034773) |
| 0.1 |
0.6 |
GO:0002087 |
regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.1 |
1.2 |
GO:0030168 |
platelet activation(GO:0030168) |
| 0.1 |
0.4 |
GO:0015868 |
purine ribonucleotide transport(GO:0015868) |
| 0.0 |
0.6 |
GO:0009143 |
nucleoside triphosphate catabolic process(GO:0009143) |
| 0.0 |
0.5 |
GO:0033211 |
adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 |
0.2 |
GO:0010808 |
positive regulation of synaptic vesicle priming(GO:0010808) positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.0 |
0.2 |
GO:0089711 |
L-glutamate transmembrane transport(GO:0089711) |
| 0.0 |
0.2 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 |
0.5 |
GO:0071803 |
positive regulation of podosome assembly(GO:0071803) |
| 0.0 |
0.3 |
GO:0002318 |
myeloid progenitor cell differentiation(GO:0002318) |
| 0.0 |
0.5 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.0 |
1.5 |
GO:0031146 |
SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 |
0.6 |
GO:0060394 |
negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 |
0.3 |
GO:0071285 |
cellular response to lithium ion(GO:0071285) |
| 0.0 |
1.5 |
GO:0045104 |
intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 |
0.6 |
GO:0001516 |
prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 |
5.0 |
GO:0007605 |
sensory perception of sound(GO:0007605) |
| 0.0 |
0.8 |
GO:2000772 |
regulation of cellular senescence(GO:2000772) |
| 0.0 |
0.1 |
GO:0035790 |
platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.0 |
0.9 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 |
0.8 |
GO:0018279 |
N-acetylneuraminate metabolic process(GO:0006054) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 |
2.1 |
GO:0016574 |
histone ubiquitination(GO:0016574) |
| 0.0 |
0.7 |
GO:0044381 |
glucose import in response to insulin stimulus(GO:0044381) regulation of glucose import in response to insulin stimulus(GO:2001273) |
| 0.0 |
0.2 |
GO:0046501 |
protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 |
0.7 |
GO:0045026 |
plasma membrane fusion(GO:0045026) |
| 0.0 |
0.1 |
GO:0071544 |
diphosphoinositol polyphosphate metabolic process(GO:0071543) diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.0 |
1.3 |
GO:0060612 |
adipose tissue development(GO:0060612) |
| 0.0 |
0.2 |
GO:0033299 |
secretion of lysosomal enzymes(GO:0033299) |
| 0.0 |
0.2 |
GO:0021902 |
commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 |
0.4 |
GO:0033129 |
positive regulation of histone phosphorylation(GO:0033129) |
| 0.0 |
0.5 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.0 |
0.8 |
GO:0010972 |
negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 |
0.2 |
GO:0031954 |
positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 |
0.1 |
GO:0006682 |
galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 |
0.1 |
GO:1900095 |
regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 |
0.3 |
GO:0060009 |
Sertoli cell development(GO:0060009) |
| 0.0 |
0.3 |
GO:0048671 |
negative regulation of collateral sprouting(GO:0048671) |
| 0.0 |
0.3 |
GO:0048144 |
fibroblast proliferation(GO:0048144) |
| 0.0 |
0.1 |
GO:0072356 |
chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.0 |
0.2 |
GO:0050860 |
negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 |
0.6 |
GO:1902476 |
chloride transmembrane transport(GO:1902476) |
| 0.0 |
0.1 |
GO:0060700 |
regulation of ribonuclease activity(GO:0060700) |
| 0.0 |
0.9 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 |
0.1 |
GO:1901837 |
negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.0 |
0.4 |
GO:0030514 |
negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 |
0.6 |
GO:0070936 |
protein K48-linked ubiquitination(GO:0070936) |
| 0.0 |
0.2 |
GO:0051044 |
positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 |
0.3 |
GO:0000413 |
protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 |
0.2 |
GO:0042744 |
hydrogen peroxide catabolic process(GO:0042744) |