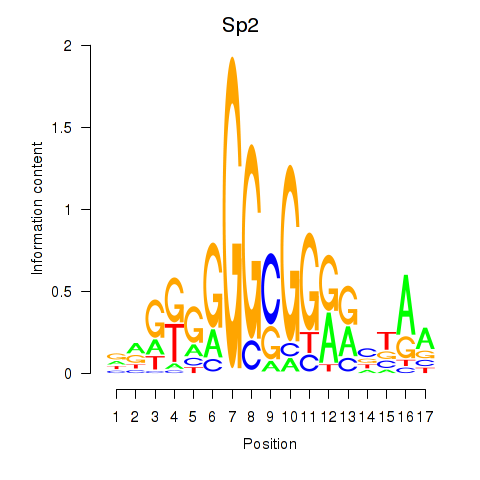
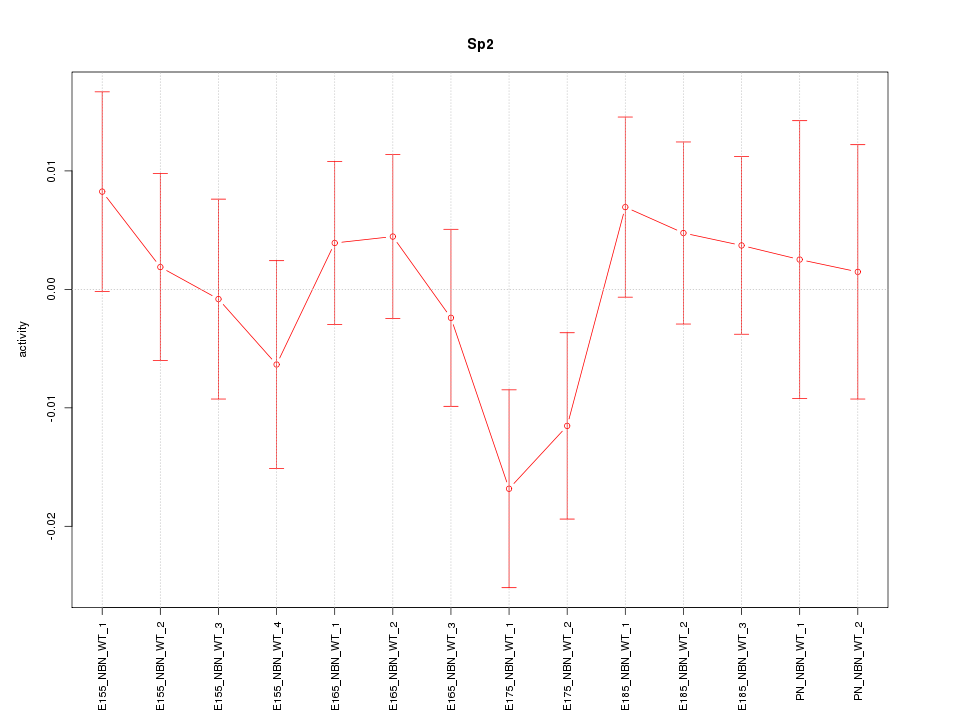
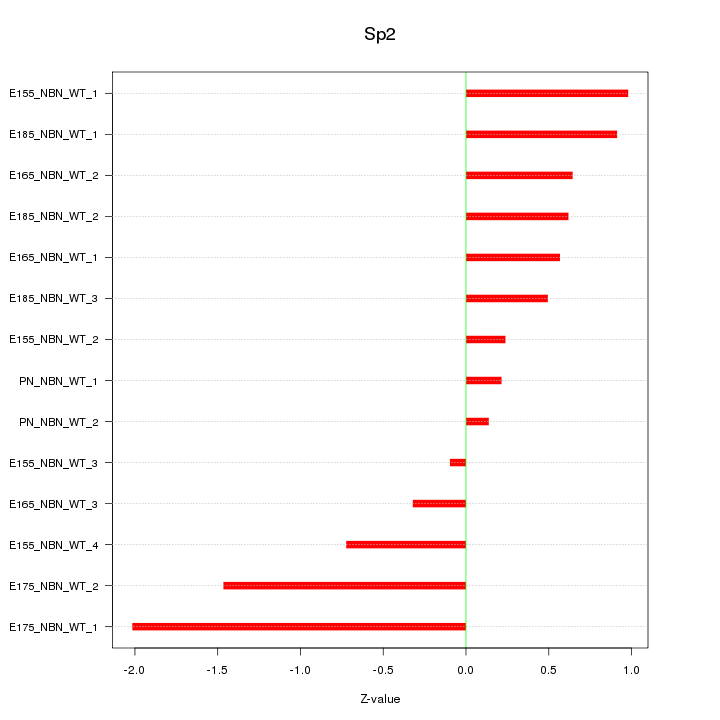
| 0.4 |
1.2 |
GO:0001869 |
regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.2 |
0.6 |
GO:1903232 |
melanosome assembly(GO:1903232) |
| 0.2 |
0.6 |
GO:0097402 |
neuroblast migration(GO:0097402) |
| 0.2 |
0.5 |
GO:0001193 |
maintenance of transcriptional fidelity during DNA-templated transcription elongation(GO:0001192) maintenance of transcriptional fidelity during DNA-templated transcription elongation from RNA polymerase II promoter(GO:0001193) |
| 0.1 |
0.4 |
GO:0019074 |
viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 |
0.8 |
GO:1904936 |
cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 |
1.6 |
GO:0002227 |
innate immune response in mucosa(GO:0002227) |
| 0.1 |
0.6 |
GO:0070345 |
negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 |
0.2 |
GO:1901228 |
positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.1 |
0.6 |
GO:0010032 |
meiotic chromosome condensation(GO:0010032) |
| 0.1 |
0.2 |
GO:0035461 |
vitamin transmembrane transport(GO:0035461) |
| 0.1 |
0.3 |
GO:1904685 |
positive regulation of metalloendopeptidase activity(GO:1904685) |
| 0.1 |
0.5 |
GO:0032792 |
negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.1 |
0.3 |
GO:0014809 |
regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 |
0.4 |
GO:0043973 |
histone H3-K4 acetylation(GO:0043973) |
| 0.1 |
1.2 |
GO:0048672 |
positive regulation of collateral sprouting(GO:0048672) |
| 0.1 |
0.3 |
GO:1901552 |
positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 |
0.3 |
GO:0007084 |
mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 |
0.3 |
GO:0007290 |
spermatid nucleus elongation(GO:0007290) |
| 0.1 |
0.3 |
GO:0061743 |
motor learning(GO:0061743) |
| 0.1 |
0.3 |
GO:0090289 |
regulation of osteoclast proliferation(GO:0090289) |
| 0.1 |
0.8 |
GO:0033280 |
response to vitamin D(GO:0033280) |
| 0.1 |
0.5 |
GO:0044829 |
positive regulation by host of viral genome replication(GO:0044829) |
| 0.1 |
0.2 |
GO:0007621 |
negative regulation of female receptivity(GO:0007621) |
| 0.1 |
0.2 |
GO:0000087 |
mitotic M phase(GO:0000087) |
| 0.1 |
0.4 |
GO:0021914 |
negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.1 |
0.4 |
GO:0036337 |
Fas signaling pathway(GO:0036337) |
| 0.1 |
0.2 |
GO:0006669 |
sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.1 |
0.3 |
GO:0071051 |
polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.1 |
0.2 |
GO:0070318 |
positive regulation of G0 to G1 transition(GO:0070318) |
| 0.1 |
0.5 |
GO:0009449 |
gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.1 |
0.5 |
GO:1902416 |
positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.1 |
0.2 |
GO:0010958 |
regulation of amino acid import(GO:0010958) regulation of L-arginine import(GO:0010963) |
| 0.1 |
0.4 |
GO:0014834 |
skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) |
| 0.1 |
0.3 |
GO:0000379 |
tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 |
0.5 |
GO:0034638 |
phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 |
0.6 |
GO:2000467 |
positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.1 |
0.4 |
GO:0048934 |
peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 |
0.2 |
GO:0090285 |
negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.1 |
0.2 |
GO:2000271 |
positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 |
0.2 |
GO:0000255 |
allantoin metabolic process(GO:0000255) |
| 0.1 |
0.2 |
GO:1903416 |
response to glycoside(GO:1903416) |
| 0.1 |
0.3 |
GO:0090292 |
nuclear migration along microfilament(GO:0031022) nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 |
0.3 |
GO:0050651 |
dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.1 |
0.5 |
GO:0051561 |
positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.1 |
0.2 |
GO:0001827 |
inner cell mass cell fate commitment(GO:0001827) |
| 0.1 |
0.2 |
GO:0034975 |
protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 |
0.3 |
GO:0001561 |
fatty acid alpha-oxidation(GO:0001561) |
| 0.1 |
0.4 |
GO:0018401 |
peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 |
0.2 |
GO:0032513 |
negative regulation of protein phosphatase type 2B activity(GO:0032513) |
| 0.1 |
0.7 |
GO:2000009 |
negative regulation of protein localization to cell surface(GO:2000009) |
| 0.1 |
0.2 |
GO:0090526 |
response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.1 |
0.6 |
GO:0000244 |
spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 |
0.2 |
GO:0042796 |
snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 |
0.2 |
GO:0006982 |
response to lipid hydroperoxide(GO:0006982) |
| 0.0 |
0.3 |
GO:0070458 |
detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.0 |
0.1 |
GO:1904706 |
negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.0 |
0.6 |
GO:0045792 |
negative regulation of cell size(GO:0045792) |
| 0.0 |
0.2 |
GO:0002741 |
positive regulation of cytokine secretion involved in immune response(GO:0002741) |
| 0.0 |
0.2 |
GO:0045054 |
constitutive secretory pathway(GO:0045054) |
| 0.0 |
0.3 |
GO:1904217 |
regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 |
0.1 |
GO:0031120 |
snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 |
0.2 |
GO:0001842 |
neural fold formation(GO:0001842) |
| 0.0 |
0.3 |
GO:0006477 |
protein sulfation(GO:0006477) |
| 0.0 |
0.6 |
GO:0036065 |
fucosylation(GO:0036065) |
| 0.0 |
0.2 |
GO:0006167 |
AMP biosynthetic process(GO:0006167) |
| 0.0 |
0.2 |
GO:1903031 |
regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 |
0.2 |
GO:0042167 |
heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 |
0.5 |
GO:0042541 |
hemoglobin biosynthetic process(GO:0042541) |
| 0.0 |
0.1 |
GO:0006421 |
asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 |
0.2 |
GO:1904690 |
regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.0 |
0.6 |
GO:0043567 |
regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 |
0.3 |
GO:0035330 |
regulation of hippo signaling(GO:0035330) |
| 0.0 |
0.6 |
GO:0006924 |
activation-induced cell death of T cells(GO:0006924) |
| 0.0 |
0.4 |
GO:0098903 |
regulation of membrane repolarization during action potential(GO:0098903) |
| 0.0 |
0.1 |
GO:0007341 |
penetration of zona pellucida(GO:0007341) positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 |
0.1 |
GO:1904749 |
regulation of protein localization to nucleolus(GO:1904749) positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.0 |
0.5 |
GO:0046697 |
decidualization(GO:0046697) |
| 0.0 |
0.7 |
GO:0000028 |
ribosomal small subunit assembly(GO:0000028) |
| 0.0 |
0.1 |
GO:1900110 |
negative regulation of histone H3-K9 dimethylation(GO:1900110) |
| 0.0 |
0.1 |
GO:1904431 |
positive regulation of t-circle formation(GO:1904431) |
| 0.0 |
0.3 |
GO:0042538 |
hyperosmotic salinity response(GO:0042538) |
| 0.0 |
0.0 |
GO:0060266 |
negative regulation of respiratory burst involved in inflammatory response(GO:0060266) negative regulation of respiratory burst(GO:0060268) |
| 0.0 |
0.1 |
GO:0010989 |
negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 |
0.1 |
GO:0070829 |
heterochromatin maintenance(GO:0070829) |
| 0.0 |
0.1 |
GO:0009301 |
snRNA transcription(GO:0009301) |
| 0.0 |
0.1 |
GO:0031652 |
positive regulation of heat generation(GO:0031652) |
| 0.0 |
0.1 |
GO:0090038 |
negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 |
0.1 |
GO:0002536 |
respiratory burst involved in inflammatory response(GO:0002536) |
| 0.0 |
0.2 |
GO:0006123 |
mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 |
0.1 |
GO:0015772 |
disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 |
0.3 |
GO:0030836 |
positive regulation of actin filament depolymerization(GO:0030836) |
| 0.0 |
0.3 |
GO:2000010 |
positive regulation of protein localization to cell surface(GO:2000010) |
| 0.0 |
0.6 |
GO:0034063 |
stress granule assembly(GO:0034063) |
| 0.0 |
0.1 |
GO:0006106 |
fumarate metabolic process(GO:0006106) |
| 0.0 |
0.4 |
GO:0007413 |
axonal fasciculation(GO:0007413) |
| 0.0 |
0.1 |
GO:0003273 |
cell migration involved in endocardial cushion formation(GO:0003273) |
| 0.0 |
0.2 |
GO:0075522 |
IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 |
0.2 |
GO:0006102 |
isocitrate metabolic process(GO:0006102) |
| 0.0 |
0.1 |
GO:0098734 |
protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 |
0.4 |
GO:0009081 |
branched-chain amino acid metabolic process(GO:0009081) |
| 0.0 |
0.1 |
GO:0032962 |
positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 |
0.1 |
GO:2000049 |
positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 |
0.3 |
GO:0035269 |
protein O-linked mannosylation(GO:0035269) |
| 0.0 |
0.4 |
GO:0006907 |
pinocytosis(GO:0006907) |
| 0.0 |
0.1 |
GO:0001771 |
immunological synapse formation(GO:0001771) |
| 0.0 |
0.1 |
GO:0060244 |
negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.0 |
0.2 |
GO:0000291 |
nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.0 |
0.6 |
GO:0050873 |
brown fat cell differentiation(GO:0050873) |
| 0.0 |
0.6 |
GO:0019884 |
antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.0 |
0.2 |
GO:0043649 |
dicarboxylic acid catabolic process(GO:0043649) |
| 0.0 |
0.1 |
GO:0031117 |
positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 |
0.1 |
GO:1901675 |
negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 |
0.1 |
GO:1903553 |
positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 |
0.1 |
GO:0071494 |
cellular response to UV-C(GO:0071494) |
| 0.0 |
0.1 |
GO:0042276 |
error-prone translesion synthesis(GO:0042276) |
| 0.0 |
0.1 |
GO:2000020 |
positive regulation of male gonad development(GO:2000020) |
| 0.0 |
0.3 |
GO:0021796 |
cerebral cortex regionalization(GO:0021796) |
| 0.0 |
0.1 |
GO:0043653 |
mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 |
0.1 |
GO:0033563 |
dorsal/ventral axon guidance(GO:0033563) |
| 0.0 |
0.4 |
GO:0016578 |
histone deubiquitination(GO:0016578) |
| 0.0 |
0.1 |
GO:0090166 |
Golgi disassembly(GO:0090166) |
| 0.0 |
0.2 |
GO:0045217 |
cell-cell junction maintenance(GO:0045217) |
| 0.0 |
0.0 |
GO:0015910 |
peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 |
0.1 |
GO:0070475 |
rRNA base methylation(GO:0070475) |
| 0.0 |
0.1 |
GO:0061179 |
negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 |
0.2 |
GO:0060736 |
prostate gland growth(GO:0060736) |
| 0.0 |
0.2 |
GO:2001241 |
positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 |
0.0 |
GO:0006556 |
S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 |
0.3 |
GO:0001522 |
pseudouridine synthesis(GO:0001522) |
| 0.0 |
0.1 |
GO:0060120 |
auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 |
0.0 |
GO:0014877 |
response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 |
0.1 |
GO:0021559 |
trigeminal nerve development(GO:0021559) |
| 0.0 |
0.3 |
GO:0010718 |
positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 |
0.2 |
GO:0031274 |
positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 |
0.3 |
GO:0010867 |
positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 |
0.2 |
GO:0007216 |
G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 |
0.1 |
GO:0042407 |
cristae formation(GO:0042407) |
| 0.0 |
0.2 |
GO:0005980 |
polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 |
0.5 |
GO:0006506 |
GPI anchor biosynthetic process(GO:0006506) |
| 0.0 |
0.3 |
GO:0008053 |
mitochondrial fusion(GO:0008053) |
| 0.0 |
0.3 |
GO:0048013 |
ephrin receptor signaling pathway(GO:0048013) |
| 0.0 |
0.1 |
GO:0097368 |
establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 |
0.1 |
GO:0032201 |
telomere maintenance via semi-conservative replication(GO:0032201) |