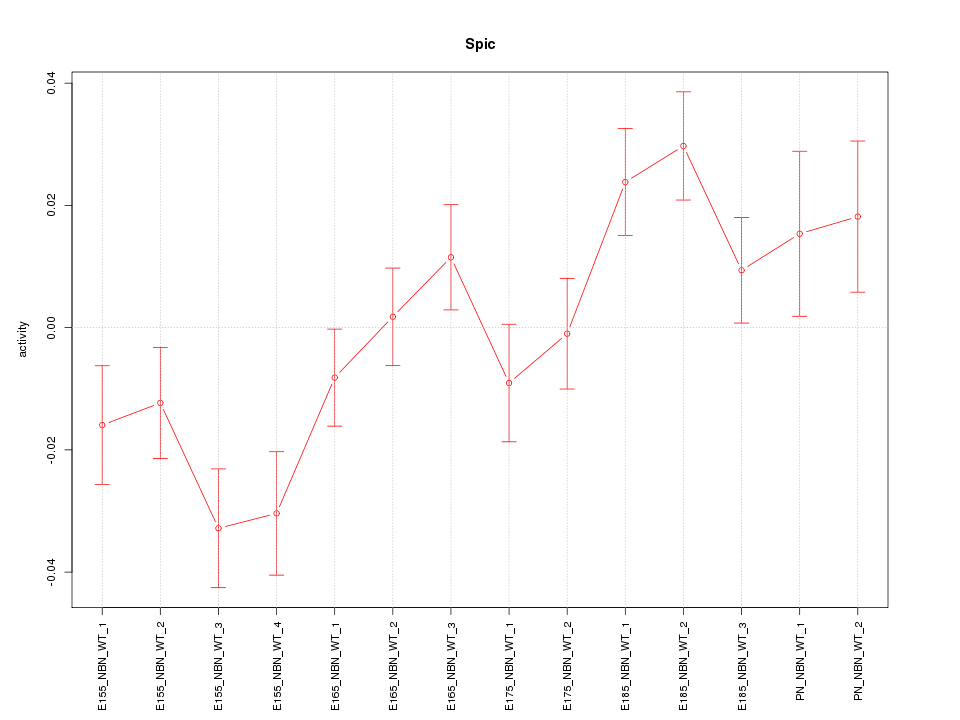
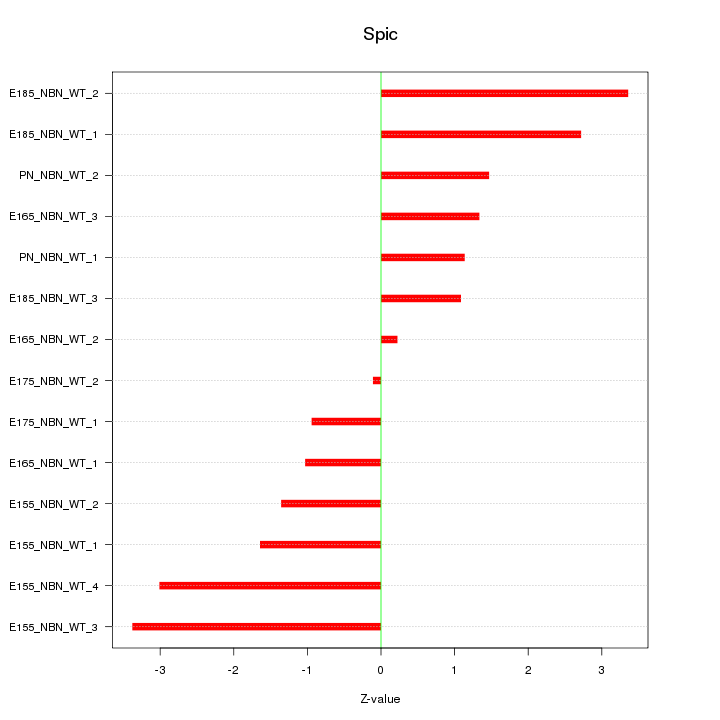
Motif ID: Spic
Z-value: 1.930
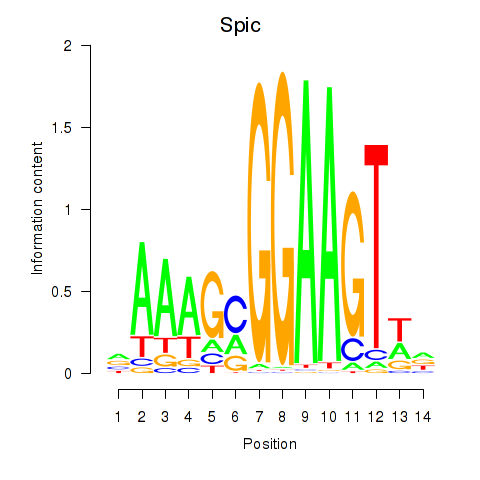
Transcription factors associated with Spic:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Spic | ENSMUSG00000004359.10 | Spic |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Spic | mm10_v2_chr10_-_88683021_88683025 | 0.16 | 6.0e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 17.3 | GO:0001803 | type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 2.5 | 9.9 | GO:1900426 | positive regulation of defense response to bacterium(GO:1900426) |
| 2.2 | 15.2 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 2.0 | 6.1 | GO:0016068 | regulation of type I hypersensitivity(GO:0001810) type I hypersensitivity(GO:0016068) |
| 1.0 | 3.1 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 1.0 | 3.1 | GO:2001187 | positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.8 | 2.4 | GO:0070476 | rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.7 | 10.6 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.6 | 1.2 | GO:0032701 | negative regulation of interleukin-18 production(GO:0032701) |
| 0.6 | 1.8 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.6 | 2.3 | GO:0006742 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.6 | 2.3 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.5 | 2.5 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.5 | 1.5 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.5 | 8.2 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.5 | 1.4 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.5 | 1.4 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.4 | 1.2 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.4 | 1.2 | GO:0045014 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.4 | 1.1 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 0.4 | 2.5 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.3 | 1.0 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) interleukin-3 production(GO:0032632) |
| 0.3 | 1.0 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.3 | 1.3 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.3 | 1.2 | GO:0050904 | diapedesis(GO:0050904) |
| 0.3 | 2.8 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.3 | 3.9 | GO:0010388 | cullin deneddylation(GO:0010388) |
| 0.3 | 4.9 | GO:0019835 | cytolysis(GO:0019835) |
| 0.3 | 0.8 | GO:1900673 | olefin metabolic process(GO:1900673) |
| 0.3 | 2.1 | GO:0071732 | cellular response to nitric oxide(GO:0071732) |
| 0.3 | 1.0 | GO:1903984 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.3 | 1.5 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.2 | 1.2 | GO:2000157 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.2 | 0.9 | GO:0003430 | tolerance induction to self antigen(GO:0002513) growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.2 | 1.8 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.2 | 0.7 | GO:0098501 | polynucleotide dephosphorylation(GO:0098501) |
| 0.2 | 1.5 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.2 | 1.2 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.2 | 3.1 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.2 | 2.1 | GO:0051126 | negative regulation of actin nucleation(GO:0051126) |
| 0.2 | 0.7 | GO:0018343 | protein farnesylation(GO:0018343) negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.2 | 1.3 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.2 | 0.2 | GO:0002309 | T cell proliferation involved in immune response(GO:0002309) |
| 0.2 | 2.1 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 0.6 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.1 | 2.3 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.1 | 0.4 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.1 | 0.4 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.1 | 1.9 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.1 | 1.4 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 0.7 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.1 | 1.9 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 0.7 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.1 | 0.6 | GO:0031052 | programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) |
| 0.1 | 0.4 | GO:0010920 | negative regulation of inositol phosphate biosynthetic process(GO:0010920) |
| 0.1 | 0.6 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.1 | 1.7 | GO:0045954 | positive regulation of natural killer cell mediated immunity(GO:0002717) positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.1 | 0.6 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.1 | 0.3 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.1 | 0.5 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.1 | 1.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.7 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) regulation of wound healing, spreading of epidermal cells(GO:1903689) positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.1 | 1.3 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 0.3 | GO:0048611 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.1 | 1.3 | GO:0018126 | protein hydroxylation(GO:0018126) |
| 0.1 | 1.6 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 1.2 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.1 | 0.5 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.1 | 0.2 | GO:0075136 | response to defenses of other organism involved in symbiotic interaction(GO:0052173) response to host defenses(GO:0052200) response to host(GO:0075136) |
| 0.1 | 0.5 | GO:0032471 | negative regulation of endoplasmic reticulum calcium ion concentration(GO:0032471) |
| 0.1 | 1.1 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.1 | 2.3 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.1 | 0.5 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.1 | 0.2 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.1 | 1.1 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.1 | 0.1 | GO:0045410 | positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 0.1 | 0.2 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 0.2 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.1 | 0.7 | GO:0015816 | glycine transport(GO:0015816) |
| 0.1 | 1.1 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 0.6 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.1 | 0.3 | GO:0010730 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.1 | 0.2 | GO:0071139 | resolution of recombination intermediates(GO:0071139) |
| 0.1 | 0.3 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.1 | 1.4 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 0.5 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.6 | GO:0050965 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 0.3 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.7 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.8 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.6 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.7 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.8 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.2 | GO:0018202 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) peptidyl-histidine modification(GO:0018202) |
| 0.0 | 0.3 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 1.0 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.4 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.5 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.4 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.0 | 0.3 | GO:0090037 | positive regulation of protein kinase C signaling(GO:0090037) |
| 0.0 | 0.6 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.3 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.1 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.2 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.0 | 0.4 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.0 | 0.5 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 4.1 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.0 | 0.1 | GO:0000087 | mitotic M phase(GO:0000087) |
| 0.0 | 0.2 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.6 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.8 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.0 | 0.7 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.7 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.0 | 0.1 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.0 | 0.0 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.2 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.5 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.3 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.8 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.1 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.4 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.7 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.1 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.0 | 0.8 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.2 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 2.6 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.7 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.3 | GO:0030201 | heparan sulfate proteoglycan metabolic process(GO:0030201) |
| 0.0 | 0.1 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.3 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.0 | 0.8 | GO:0035904 | aorta development(GO:0035904) |
| 0.0 | 0.6 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 0.2 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.4 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.3 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.6 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.4 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.8 | 3.9 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.5 | 2.6 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.3 | 24.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.3 | 8.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 0.7 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.2 | 3.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 1.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.2 | 1.3 | GO:0000800 | lateral element(GO:0000800) |
| 0.2 | 3.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.2 | 0.8 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.2 | 1.5 | GO:0005818 | aster(GO:0005818) |
| 0.2 | 0.7 | GO:0032021 | NELF complex(GO:0032021) |
| 0.2 | 1.5 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.1 | 2.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 1.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.5 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.1 | 4.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 1.5 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.1 | 0.5 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 23.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 0.6 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 3.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 0.4 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 0.6 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 0.3 | GO:0097651 | phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.1 | 1.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 0.4 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.1 | 0.8 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.5 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 1.9 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.4 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.1 | 2.2 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 1.8 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.6 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 0.8 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 23.5 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.1 | 0.3 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 2.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.4 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.2 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.0 | 0.3 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.6 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.5 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 2.6 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.5 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.3 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 1.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.3 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.4 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 1.2 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.3 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.4 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.0 | 0.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.0 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.5 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 2.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.8 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.7 | GO:0005604 | basement membrane(GO:0005604) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 23.4 | GO:0019864 | IgG binding(GO:0019864) |
| 2.0 | 8.0 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 1.0 | 5.8 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.7 | 2.2 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.6 | 0.6 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.5 | 4.4 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.5 | 2.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.5 | 1.4 | GO:0072541 | peroxynitrite reductase activity(GO:0072541) |
| 0.4 | 3.1 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.4 | 1.3 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.4 | 1.6 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.4 | 2.5 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.3 | 1.0 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.3 | 1.5 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.3 | 7.6 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.3 | 0.8 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.3 | 1.4 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.2 | 0.7 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.2 | 0.9 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.2 | 1.2 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.2 | 3.6 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.2 | 1.3 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.2 | 0.5 | GO:0004368 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) |
| 0.2 | 1.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.2 | 1.4 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 0.7 | GO:0046403 | polynucleotide 3'-phosphatase activity(GO:0046403) |
| 0.1 | 1.9 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 1.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.5 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 1.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 1.9 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 1.0 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 1.4 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 1.0 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.1 | 0.5 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 1.0 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.1 | 2.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.5 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.1 | 0.7 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 0.7 | GO:0052872 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) phenanthrene 9,10-monooxygenase activity(GO:0018636) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) tocotrienol omega-hydroxylase activity(GO:0052872) thalianol hydroxylase activity(GO:0080014) |
| 0.1 | 0.3 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.1 | 2.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.2 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 1.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.2 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 1.2 | GO:0035380 | C-3 sterol dehydrogenase (C-4 sterol decarboxylase) activity(GO:0000252) mevaldate reductase activity(GO:0004495) gluconate dehydrogenase activity(GO:0008875) epoxide dehydrogenase activity(GO:0018451) 5-exo-hydroxycamphor dehydrogenase activity(GO:0018452) 2-hydroxytetrahydrofuran dehydrogenase activity(GO:0018453) acetoin dehydrogenase activity(GO:0019152) phenylcoumaran benzylic ether reductase activity(GO:0032442) D-xylose:NADP reductase activity(GO:0032866) L-arabinose:NADP reductase activity(GO:0032867) D-arabinitol dehydrogenase, D-ribulose forming (NADP+) activity(GO:0033709) (R)-(-)-1,2,3,4-tetrahydronaphthol dehydrogenase activity(GO:0034831) 3-hydroxymenthone dehydrogenase activity(GO:0034840) very long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0035380) dihydrotestosterone 17-beta-dehydrogenase activity(GO:0035410) (R)-2-hydroxyisocaproate dehydrogenase activity(GO:0043713) L-arabinose 1-dehydrogenase (NADP+) activity(GO:0044103) L-xylulose reductase (NAD+) activity(GO:0044105) 3-ketoglucose-reductase activity(GO:0048258) D-arabinitol dehydrogenase, D-xylulose forming (NADP+) activity(GO:0052677) |
| 0.1 | 0.2 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 1.5 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.4 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.3 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.8 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.0 | 7.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.5 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.0 | 0.4 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.2 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.4 | GO:0016248 | ion channel inhibitor activity(GO:0008200) channel inhibitor activity(GO:0016248) |
| 0.0 | 0.9 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.9 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 1.9 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.6 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 2.0 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 1.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.3 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.4 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.6 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.3 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.4 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 2.1 | GO:0019209 | kinase activator activity(GO:0019209) |
| 0.0 | 1.2 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.5 | GO:0044653 | trehalase activity(GO:0015927) dextrin alpha-glucosidase activity(GO:0044653) starch alpha-glucosidase activity(GO:0044654) beta-glucanase activity(GO:0052736) beta-6-sulfate-N-acetylglucosaminidase activity(GO:0052769) glucan endo-1,4-beta-glucosidase activity(GO:0052859) |
| 0.0 | 0.3 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.1 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 1.0 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 2.0 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.0 | GO:0034952 | 3-octaprenyl-4-hydroxybenzoate carboxy-lyase activity(GO:0008694) 2-hydroxy-3-carboxy-6-oxo-7-methylocta-2,4-dienoate decarboxylase activity(GO:0018791) bis(4-chlorophenyl)acetate decarboxylase activity(GO:0018792) 3,5-dibromo-4-hydroxybenzoate decarboxylase activity(GO:0018793) 2-hydroxyisobutyrate decarboxylase activity(GO:0018794) 2-hydroxy-2-methyl-1,3-dicarbonate decarboxylase activity(GO:0018795) 2-hydroxyisophthalate decarboxylase activity(GO:0034524) dimethylmalonate decarboxylase activity(GO:0034782) 2,4,4-trimethyl-3-oxopentanoate decarboxylase activity(GO:0034853) 4,4-dimethyl-3-oxopentanoate decarboxylase activity(GO:0034854) 2,3,6-trihydroxyisonicotinate decarboxylase activity(GO:0034879) phenanthrene-4,5-dicarboxylate decarboxylase activity(GO:0034923) pyrrole-2-carboxylate decarboxylase activity(GO:0034941) terephthalate decarboxylase activity(GO:0034947) malonate semialdehyde decarboxylase activity(GO:0034952) 5-amino-4-imidazole carboxylate lyase activity(GO:0043727) 2-keto-4-methylthiobutyrate aminotransferase activity(GO:0043728) 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase activity(GO:0051997) |
| 0.0 | 0.0 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 0.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 1.7 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 0.4 | GO:0017091 | AU-rich element binding(GO:0017091) |