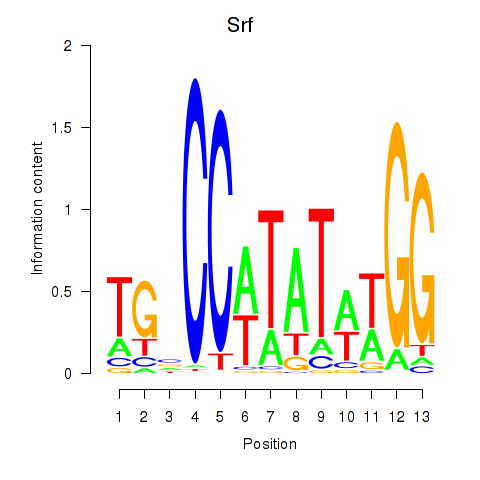
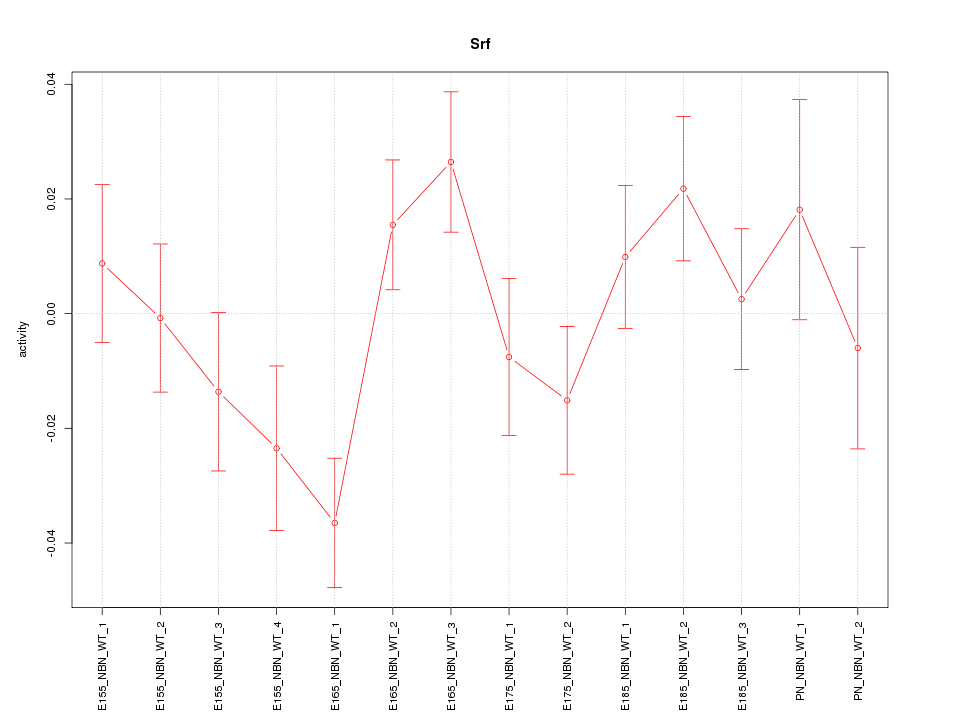
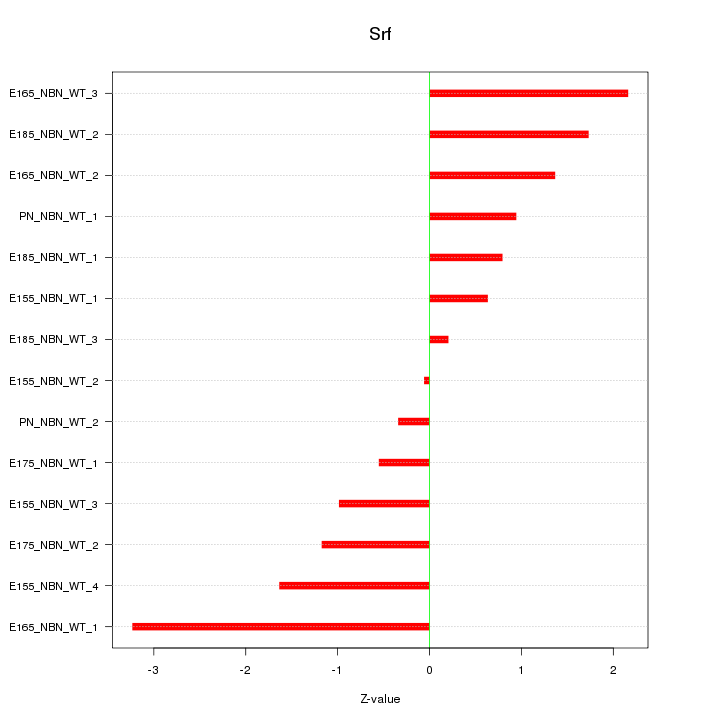
| 4.9 |
14.8 |
GO:0072223 |
metanephric glomerular mesangium development(GO:0072223) metanephric glomerular mesangial cell proliferation involved in metanephros development(GO:0072262) |
| 1.0 |
8.2 |
GO:0038028 |
insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.3 |
2.5 |
GO:1902166 |
negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.3 |
0.8 |
GO:0006116 |
NADH oxidation(GO:0006116) |
| 0.2 |
1.2 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.2 |
1.1 |
GO:0021593 |
rhombomere morphogenesis(GO:0021593) |
| 0.2 |
1.1 |
GO:0072257 |
metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.1 |
2.5 |
GO:0050765 |
negative regulation of phagocytosis(GO:0050765) |
| 0.1 |
1.0 |
GO:0030579 |
ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 |
2.5 |
GO:0035994 |
response to muscle stretch(GO:0035994) |
| 0.1 |
0.8 |
GO:1904715 |
negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.1 |
0.8 |
GO:0021797 |
forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 |
1.6 |
GO:1903504 |
regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 |
0.4 |
GO:0045358 |
negative regulation of interferon-beta biosynthetic process(GO:0045358) |
| 0.1 |
0.8 |
GO:0006488 |
dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 |
0.1 |
GO:0035793 |
negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) cell migration involved in metanephros development(GO:0035788) metanephric mesenchymal cell migration(GO:0035789) positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) regulation of metanephric mesenchymal cell migration(GO:2000589) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.1 |
1.1 |
GO:0090136 |
epithelial cell-cell adhesion(GO:0090136) |
| 0.1 |
2.6 |
GO:0071277 |
cellular response to calcium ion(GO:0071277) |
| 0.0 |
0.3 |
GO:0042535 |
positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.0 |
2.9 |
GO:0070527 |
platelet aggregation(GO:0070527) |
| 0.0 |
1.6 |
GO:0001706 |
endoderm formation(GO:0001706) |
| 0.0 |
0.5 |
GO:0050965 |
detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 |
0.1 |
GO:0048739 |
cardiac muscle fiber development(GO:0048739) |
| 0.0 |
0.2 |
GO:0006450 |
regulation of translational fidelity(GO:0006450) |
| 0.0 |
0.7 |
GO:1901522 |
positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 |
1.1 |
GO:2000179 |
positive regulation of neural precursor cell proliferation(GO:2000179) |
| 0.0 |
0.1 |
GO:0046294 |
formaldehyde catabolic process(GO:0046294) |
| 0.0 |
1.0 |
GO:2000134 |
negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 |
0.1 |
GO:0014887 |
muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 |
0.4 |
GO:0051898 |
negative regulation of protein kinase B signaling(GO:0051898) |