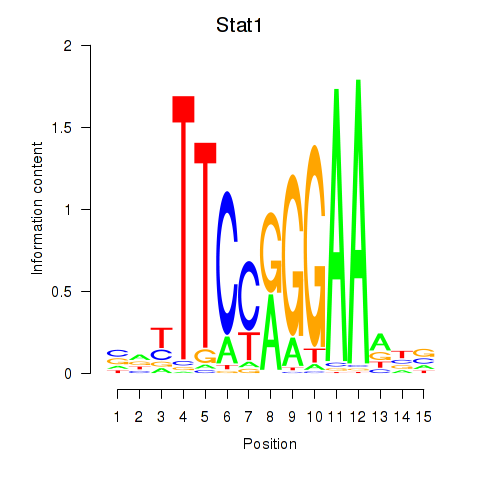
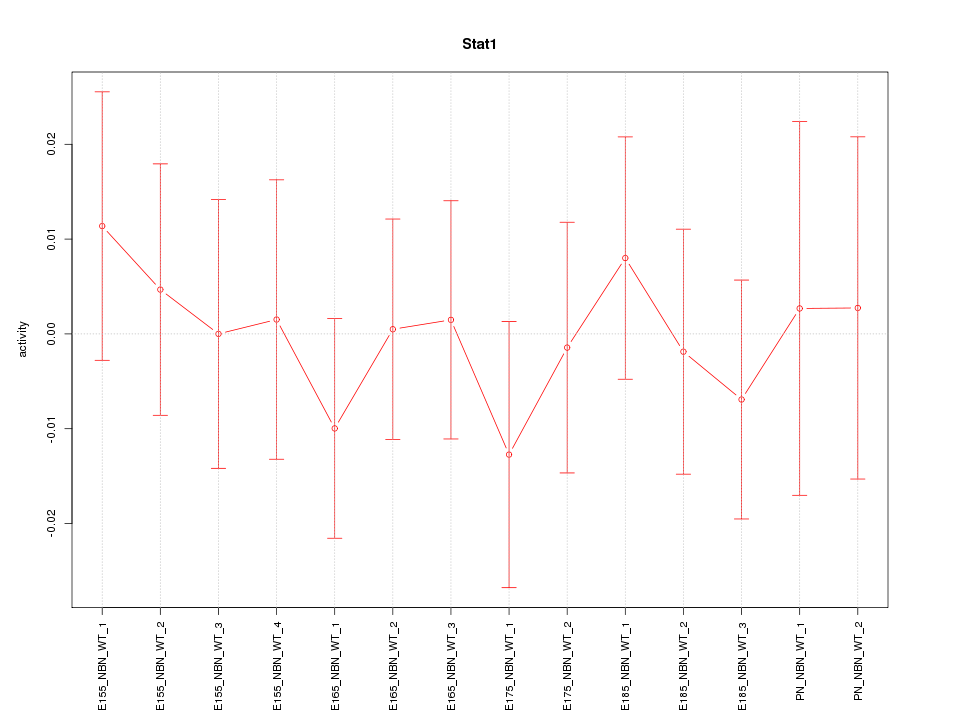
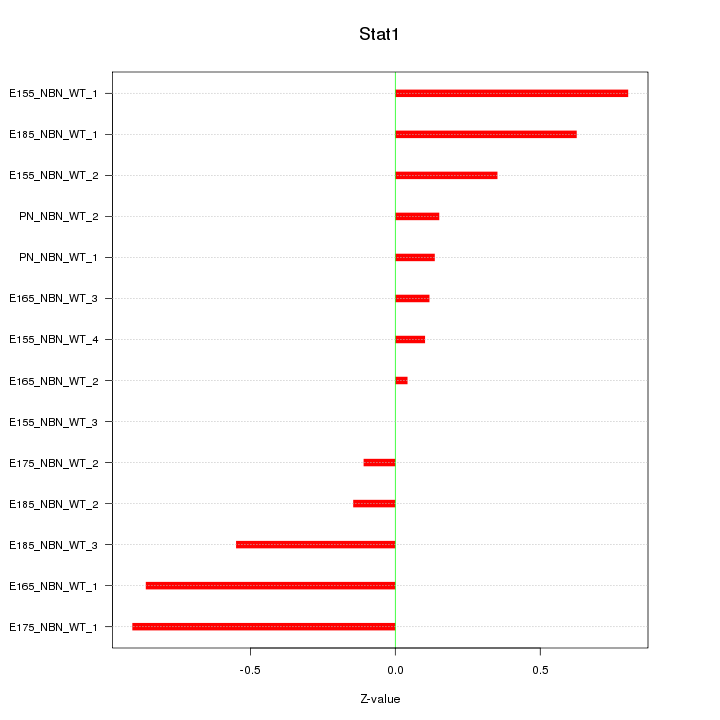
| 0.3 |
0.8 |
GO:0051892 |
negative regulation of cardioblast differentiation(GO:0051892) |
| 0.3 |
0.8 |
GO:0001869 |
regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.2 |
0.8 |
GO:0051708 |
intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) cytosol to ER transport(GO:0046967) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.2 |
0.5 |
GO:1901097 |
negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 |
0.5 |
GO:0001999 |
renal response to blood flow involved in circulatory renin-angiotensin regulation of systemic arterial blood pressure(GO:0001999) |
| 0.1 |
1.2 |
GO:0048251 |
elastic fiber assembly(GO:0048251) |
| 0.1 |
1.2 |
GO:0097421 |
liver regeneration(GO:0097421) |
| 0.1 |
0.4 |
GO:0019372 |
lipoxygenase pathway(GO:0019372) |
| 0.1 |
0.3 |
GO:2000253 |
positive regulation of feeding behavior(GO:2000253) |
| 0.1 |
0.3 |
GO:0030643 |
cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 |
0.4 |
GO:0034638 |
phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 |
0.6 |
GO:0019262 |
N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 |
0.4 |
GO:2000194 |
regulation of female gonad development(GO:2000194) |
| 0.0 |
0.1 |
GO:0060489 |
establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.0 |
0.2 |
GO:0030259 |
lipid glycosylation(GO:0030259) |
| 0.0 |
0.3 |
GO:0042730 |
fibrinolysis(GO:0042730) |
| 0.0 |
0.5 |
GO:0071378 |
growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 |
0.2 |
GO:0060576 |
intestinal epithelial cell development(GO:0060576) |
| 0.0 |
0.1 |
GO:0070682 |
proteasome regulatory particle assembly(GO:0070682) |
| 0.0 |
0.1 |
GO:0045819 |
positive regulation of glycogen catabolic process(GO:0045819) |
| 0.0 |
0.1 |
GO:0090399 |
replicative senescence(GO:0090399) |
| 0.0 |
0.2 |
GO:0007406 |
negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 |
0.5 |
GO:0009409 |
response to cold(GO:0009409) |
| 0.0 |
0.1 |
GO:0006624 |
vacuolar protein processing(GO:0006624) |
| 0.0 |
0.2 |
GO:0000722 |
telomere maintenance via recombination(GO:0000722) |
| 0.0 |
0.5 |
GO:1902230 |
negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 |
0.3 |
GO:0035235 |
ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 |
0.1 |
GO:1903361 |
protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 |
0.1 |
GO:0036120 |
cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.0 |
0.0 |
GO:1902774 |
late endosome to lysosome transport(GO:1902774) |
| 0.0 |
0.2 |
GO:2000144 |
positive regulation of DNA-templated transcription, initiation(GO:2000144) |