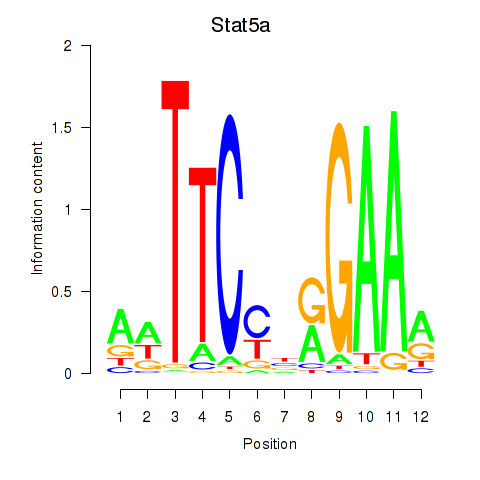
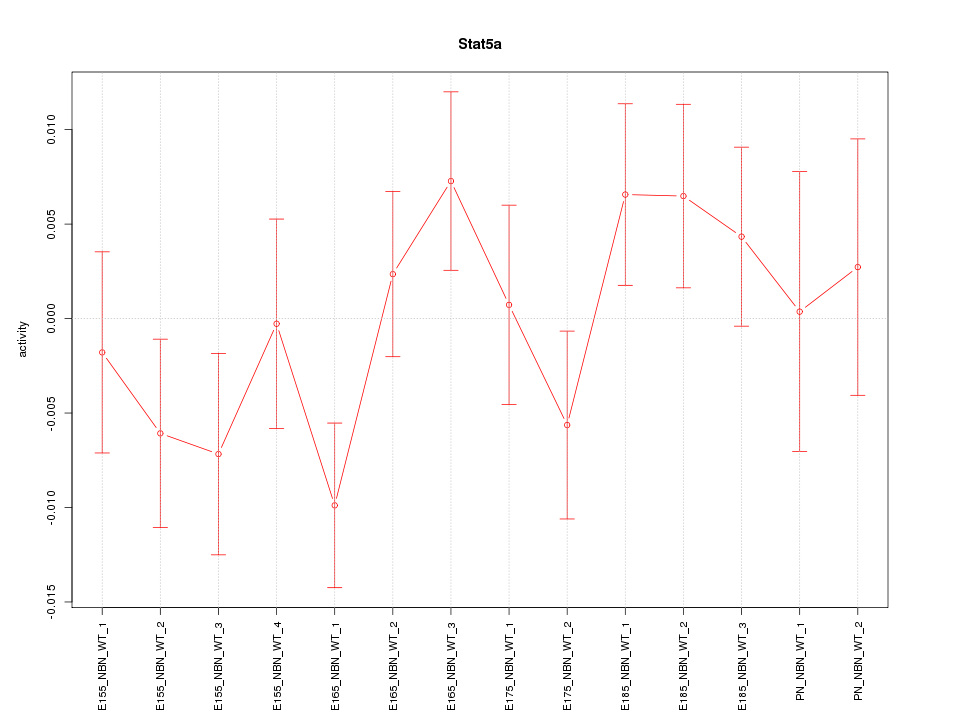
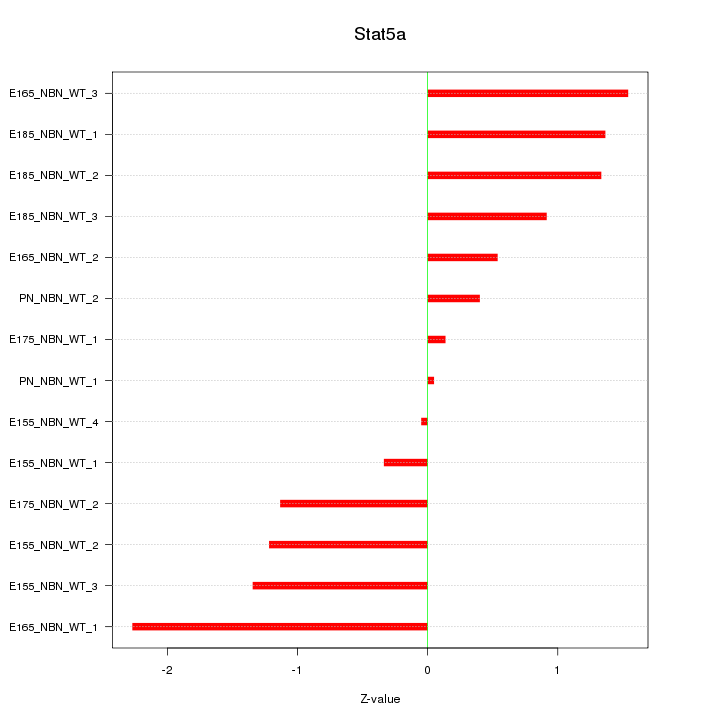
| 0.4 |
1.6 |
GO:0014012 |
peripheral nervous system axon regeneration(GO:0014012) |
| 0.4 |
1.8 |
GO:2000298 |
regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.3 |
1.4 |
GO:1901252 |
regulation of intracellular transport of viral material(GO:1901252) |
| 0.3 |
0.9 |
GO:2000402 |
negative regulation of lymphocyte migration(GO:2000402) |
| 0.3 |
0.8 |
GO:0042560 |
10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.3 |
1.4 |
GO:0070327 |
thyroid hormone transport(GO:0070327) |
| 0.3 |
0.8 |
GO:0034162 |
toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.2 |
0.7 |
GO:0032430 |
positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.2 |
0.7 |
GO:0032687 |
negative regulation of interferon-alpha production(GO:0032687) interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) negative regulation of interferon-beta biosynthetic process(GO:0045358) |
| 0.2 |
1.0 |
GO:0071726 |
response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) |
| 0.2 |
0.6 |
GO:1904706 |
response to cisplatin(GO:0072718) negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.2 |
0.6 |
GO:0060166 |
olfactory pit development(GO:0060166) |
| 0.2 |
0.6 |
GO:0001951 |
intestinal D-glucose absorption(GO:0001951) terminal web assembly(GO:1902896) |
| 0.2 |
1.3 |
GO:0034638 |
phosphatidylcholine catabolic process(GO:0034638) |
| 0.2 |
1.1 |
GO:0036072 |
intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.2 |
0.5 |
GO:0014826 |
vein smooth muscle contraction(GO:0014826) |
| 0.2 |
0.5 |
GO:0010911 |
regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.2 |
1.2 |
GO:0006363 |
termination of RNA polymerase I transcription(GO:0006363) |
| 0.2 |
0.5 |
GO:0035633 |
maintenance of blood-brain barrier(GO:0035633) |
| 0.2 |
0.5 |
GO:0002606 |
positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.2 |
0.3 |
GO:1900041 |
negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.2 |
0.5 |
GO:0002034 |
angiotensin mediated vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001998) regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 0.2 |
0.5 |
GO:0060708 |
spongiotrophoblast differentiation(GO:0060708) |
| 0.1 |
0.6 |
GO:0047484 |
regulation of response to osmotic stress(GO:0047484) |
| 0.1 |
1.3 |
GO:0097421 |
liver regeneration(GO:0097421) |
| 0.1 |
0.7 |
GO:1903012 |
positive regulation of bone development(GO:1903012) |
| 0.1 |
0.3 |
GO:0034145 |
positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.1 |
2.3 |
GO:0006958 |
complement activation, classical pathway(GO:0006958) |
| 0.1 |
0.8 |
GO:0036233 |
glycine import(GO:0036233) |
| 0.1 |
0.4 |
GO:0001869 |
regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.1 |
0.6 |
GO:0032224 |
positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.1 |
0.9 |
GO:0042730 |
fibrinolysis(GO:0042730) |
| 0.1 |
0.2 |
GO:0032701 |
negative regulation of interleukin-18 production(GO:0032701) |
| 0.1 |
0.2 |
GO:0097527 |
necroptotic signaling pathway(GO:0097527) |
| 0.1 |
0.8 |
GO:0010273 |
detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.1 |
1.2 |
GO:0033280 |
response to vitamin D(GO:0033280) |
| 0.1 |
2.4 |
GO:0050829 |
defense response to Gram-negative bacterium(GO:0050829) |
| 0.1 |
0.5 |
GO:0042360 |
vitamin E metabolic process(GO:0042360) |
| 0.1 |
0.8 |
GO:0060923 |
cardiac muscle cell fate commitment(GO:0060923) |
| 0.1 |
0.3 |
GO:0030827 |
negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.1 |
0.4 |
GO:2000383 |
regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.1 |
0.4 |
GO:0043490 |
malate-aspartate shuttle(GO:0043490) |
| 0.1 |
1.0 |
GO:0046688 |
response to copper ion(GO:0046688) |
| 0.1 |
2.2 |
GO:0010614 |
negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.1 |
0.4 |
GO:0046552 |
eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.1 |
0.7 |
GO:0071732 |
cellular response to nitric oxide(GO:0071732) |
| 0.1 |
0.4 |
GO:0001907 |
killing by symbiont of host cells(GO:0001907) induction of programmed cell death(GO:0012502) disruption by symbiont of host cell(GO:0044004) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) positive regulation of apoptotic process by virus(GO:0060139) apoptotic process involved in embryonic digit morphogenesis(GO:1902263) positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 |
0.5 |
GO:1904995 |
negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.1 |
0.9 |
GO:2000194 |
regulation of female gonad development(GO:2000194) |
| 0.1 |
0.1 |
GO:0046101 |
hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.1 |
0.8 |
GO:0044130 |
negative regulation of growth of symbiont in host(GO:0044130) negative regulation of growth of symbiont involved in interaction with host(GO:0044146) |
| 0.1 |
0.4 |
GO:0071449 |
cellular response to lipid hydroperoxide(GO:0071449) |
| 0.1 |
0.3 |
GO:0009177 |
deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) |
| 0.1 |
2.3 |
GO:0001913 |
T cell mediated cytotoxicity(GO:0001913) |
| 0.1 |
0.3 |
GO:0002430 |
complement receptor mediated signaling pathway(GO:0002430) |
| 0.1 |
0.8 |
GO:0002118 |
aggressive behavior(GO:0002118) |
| 0.1 |
0.3 |
GO:0043181 |
vacuolar sequestering(GO:0043181) |
| 0.1 |
0.3 |
GO:0086017 |
Purkinje myocyte action potential(GO:0086017) |
| 0.1 |
0.4 |
GO:2000255 |
negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 |
0.3 |
GO:0048014 |
Tie signaling pathway(GO:0048014) |
| 0.1 |
0.2 |
GO:0010752 |
regulation of cGMP-mediated signaling(GO:0010752) |
| 0.1 |
0.2 |
GO:0033566 |
gamma-tubulin complex localization(GO:0033566) |
| 0.1 |
0.2 |
GO:0072683 |
T cell extravasation(GO:0072683) |
| 0.1 |
0.3 |
GO:0007217 |
tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 |
0.6 |
GO:0071494 |
cellular response to UV-C(GO:0071494) |
| 0.1 |
0.2 |
GO:0044650 |
virion attachment to host cell(GO:0019062) adhesion of symbiont to host cell(GO:0044650) |
| 0.1 |
0.2 |
GO:0015712 |
hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 |
0.2 |
GO:0010896 |
regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.1 |
0.3 |
GO:1904425 |
negative regulation of GTP binding(GO:1904425) |
| 0.1 |
0.3 |
GO:0038145 |
macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.1 |
0.2 |
GO:0071603 |
endothelial cell-cell adhesion(GO:0071603) |
| 0.1 |
0.2 |
GO:0018298 |
protein-chromophore linkage(GO:0018298) |
| 0.1 |
0.5 |
GO:0016139 |
glycoside catabolic process(GO:0016139) |
| 0.1 |
0.2 |
GO:0040001 |
establishment of mitotic spindle localization(GO:0040001) |
| 0.1 |
0.1 |
GO:0021529 |
spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.1 |
0.2 |
GO:0060729 |
intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 |
0.3 |
GO:0021914 |
negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.1 |
0.4 |
GO:0001955 |
blood vessel maturation(GO:0001955) |
| 0.1 |
0.3 |
GO:0014835 |
myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.1 |
0.2 |
GO:0046013 |
T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 |
0.5 |
GO:2001214 |
positive regulation of vasculogenesis(GO:2001214) |
| 0.1 |
0.3 |
GO:2000680 |
regulation of rubidium ion transport(GO:2000680) |
| 0.1 |
0.3 |
GO:0050747 |
positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.1 |
0.4 |
GO:0001302 |
replicative cell aging(GO:0001302) |
| 0.1 |
0.2 |
GO:0006166 |
purine ribonucleoside salvage(GO:0006166) adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.1 |
0.1 |
GO:0001866 |
NK T cell proliferation(GO:0001866) |
| 0.1 |
1.3 |
GO:0070208 |
protein heterotrimerization(GO:0070208) |
| 0.1 |
0.5 |
GO:0006729 |
tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 |
0.2 |
GO:0035552 |
oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.1 |
0.2 |
GO:1904274 |
tricellular tight junction assembly(GO:1904274) |
| 0.1 |
0.4 |
GO:0097460 |
ferrous iron import into cell(GO:0097460) |
| 0.1 |
0.5 |
GO:0015695 |
organic cation transport(GO:0015695) |
| 0.1 |
0.6 |
GO:0043252 |
sodium-independent organic anion transport(GO:0043252) |
| 0.1 |
0.3 |
GO:0032725 |
positive regulation of granulocyte macrophage colony-stimulating factor production(GO:0032725) |
| 0.1 |
0.4 |
GO:0038028 |
insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.1 |
0.4 |
GO:0006977 |
DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.1 |
0.2 |
GO:0045041 |
protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 |
0.7 |
GO:0032757 |
positive regulation of interleukin-8 production(GO:0032757) |
| 0.1 |
1.6 |
GO:0071353 |
cellular response to interleukin-4(GO:0071353) |
| 0.1 |
0.4 |
GO:0015862 |
uridine transport(GO:0015862) |
| 0.1 |
0.8 |
GO:0090136 |
epithelial cell-cell adhesion(GO:0090136) |
| 0.1 |
0.6 |
GO:0019262 |
N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 |
0.5 |
GO:0033631 |
cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 |
0.1 |
GO:0097461 |
ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) |
| 0.0 |
0.2 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.0 |
0.2 |
GO:2000323 |
progesterone receptor signaling pathway(GO:0050847) negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 |
0.1 |
GO:0015960 |
diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.0 |
0.2 |
GO:0010756 |
positive regulation of plasminogen activation(GO:0010756) |
| 0.0 |
0.2 |
GO:0006624 |
vacuolar protein processing(GO:0006624) |
| 0.0 |
0.5 |
GO:0090385 |
phagosome-lysosome fusion(GO:0090385) |
| 0.0 |
0.2 |
GO:0010920 |
negative regulation of inositol phosphate biosynthetic process(GO:0010920) positive regulation of integrin activation(GO:0033625) |
| 0.0 |
0.6 |
GO:0061299 |
retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 |
0.1 |
GO:0003096 |
renal sodium ion transport(GO:0003096) |
| 0.0 |
0.1 |
GO:0070602 |
regulation of centromeric sister chromatid cohesion(GO:0070602) positive regulation of determination of dorsal identity(GO:2000017) |
| 0.0 |
0.2 |
GO:0070236 |
negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 |
0.2 |
GO:2000253 |
positive regulation of feeding behavior(GO:2000253) |
| 0.0 |
0.3 |
GO:0003383 |
apical constriction(GO:0003383) |
| 0.0 |
0.1 |
GO:0002924 |
negative regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002924) |
| 0.0 |
0.1 |
GO:0032066 |
nucleolus to nucleoplasm transport(GO:0032066) |
| 0.0 |
0.3 |
GO:0006685 |
sphingomyelin catabolic process(GO:0006685) |
| 0.0 |
0.3 |
GO:0044027 |
hypermethylation of CpG island(GO:0044027) |
| 0.0 |
0.2 |
GO:0009414 |
response to water deprivation(GO:0009414) |
| 0.0 |
0.1 |
GO:0035106 |
operant conditioning(GO:0035106) |
| 0.0 |
0.1 |
GO:0002710 |
negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 |
0.2 |
GO:0036151 |
phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.0 |
0.9 |
GO:0045047 |
protein targeting to ER(GO:0045047) |
| 0.0 |
0.1 |
GO:2001245 |
regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 |
0.2 |
GO:0018002 |
N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 |
0.3 |
GO:0016480 |
negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 |
0.2 |
GO:1903553 |
positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 |
0.0 |
GO:0043388 |
positive regulation of DNA binding(GO:0043388) regulation of DNA binding(GO:0051101) |
| 0.0 |
0.2 |
GO:0034316 |
negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) negative regulation of actin nucleation(GO:0051126) |
| 0.0 |
0.3 |
GO:0071361 |
cellular response to ethanol(GO:0071361) |
| 0.0 |
0.2 |
GO:0046339 |
diacylglycerol metabolic process(GO:0046339) |
| 0.0 |
0.2 |
GO:0003406 |
retinal pigment epithelium development(GO:0003406) |
| 0.0 |
0.2 |
GO:1903690 |
negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 |
0.1 |
GO:2000048 |
negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 |
0.3 |
GO:0090050 |
positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 |
0.4 |
GO:1902947 |
regulation of tau-protein kinase activity(GO:1902947) |
| 0.0 |
0.5 |
GO:0034501 |
protein localization to kinetochore(GO:0034501) |
| 0.0 |
0.2 |
GO:0045200 |
establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 |
0.2 |
GO:0036462 |
TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 |
0.1 |
GO:0097211 |
response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 |
0.2 |
GO:0006172 |
ADP biosynthetic process(GO:0006172) |
| 0.0 |
0.2 |
GO:0036089 |
cleavage furrow formation(GO:0036089) |
| 0.0 |
0.2 |
GO:0010032 |
meiotic chromosome condensation(GO:0010032) |
| 0.0 |
0.2 |
GO:1904721 |
regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 |
0.1 |
GO:0002865 |
negative regulation of acute inflammatory response to antigenic stimulus(GO:0002865) |
| 0.0 |
0.2 |
GO:0070171 |
negative regulation of tooth mineralization(GO:0070171) |
| 0.0 |
0.2 |
GO:0031584 |
activation of phospholipase D activity(GO:0031584) |
| 0.0 |
0.1 |
GO:0019065 |
receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 |
0.2 |
GO:2001170 |
negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.0 |
0.2 |
GO:0032261 |
purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.0 |
0.1 |
GO:0045204 |
MAPK export from nucleus(GO:0045204) |
| 0.0 |
0.2 |
GO:0051152 |
positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 |
0.4 |
GO:0044458 |
motile cilium assembly(GO:0044458) |
| 0.0 |
0.1 |
GO:0050812 |
regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 |
0.1 |
GO:0036518 |
chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.0 |
0.2 |
GO:0003215 |
cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 |
1.0 |
GO:0001937 |
negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.0 |
0.1 |
GO:2000210 |
positive regulation of anoikis(GO:2000210) |
| 0.0 |
0.1 |
GO:0021508 |
floor plate formation(GO:0021508) floor plate morphogenesis(GO:0033505) |
| 0.0 |
0.2 |
GO:0033623 |
regulation of integrin activation(GO:0033623) |
| 0.0 |
0.6 |
GO:0033006 |
regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.0 |
0.4 |
GO:0010574 |
regulation of vascular endothelial growth factor production(GO:0010574) |
| 0.0 |
0.2 |
GO:0007021 |
tubulin complex assembly(GO:0007021) |
| 0.0 |
0.1 |
GO:0035372 |
protein localization to microtubule(GO:0035372) |
| 0.0 |
0.1 |
GO:0017183 |
peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 |
0.4 |
GO:0042759 |
long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 |
0.1 |
GO:0044849 |
estrous cycle(GO:0044849) |
| 0.0 |
0.3 |
GO:0032291 |
central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 |
0.2 |
GO:0071763 |
nuclear membrane organization(GO:0071763) |
| 0.0 |
0.1 |
GO:0006369 |
termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 |
0.1 |
GO:1904729 |
regulation of intestinal cholesterol absorption(GO:0030300) regulation of intestinal absorption(GO:1904478) regulation of intestinal lipid absorption(GO:1904729) |
| 0.0 |
0.1 |
GO:0035021 |
negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 |
0.2 |
GO:1903352 |
ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.0 |
0.1 |
GO:0006478 |
peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 |
0.1 |
GO:0019478 |
D-amino acid catabolic process(GO:0019478) |
| 0.0 |
0.3 |
GO:0019368 |
fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 |
0.1 |
GO:0045039 |
protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 |
0.1 |
GO:0002036 |
regulation of L-glutamate transport(GO:0002036) |
| 0.0 |
0.4 |
GO:0045063 |
T-helper 1 cell differentiation(GO:0045063) |
| 0.0 |
0.0 |
GO:0006680 |
glucosylceramide catabolic process(GO:0006680) |
| 0.0 |
0.2 |
GO:0030432 |
peristalsis(GO:0030432) |
| 0.0 |
0.1 |
GO:0098700 |
aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 |
0.2 |
GO:0006527 |
arginine catabolic process(GO:0006527) |
| 0.0 |
0.4 |
GO:0046597 |
negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 |
0.1 |
GO:1904211 |
membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.0 |
0.1 |
GO:1903772 |
regulation of viral budding via host ESCRT complex(GO:1903772) |
| 0.0 |
0.3 |
GO:0032525 |
somite rostral/caudal axis specification(GO:0032525) |
| 0.0 |
0.1 |
GO:0032534 |
regulation of microvillus assembly(GO:0032534) |
| 0.0 |
0.0 |
GO:0010566 |
regulation of ketone biosynthetic process(GO:0010566) |
| 0.0 |
0.1 |
GO:0019236 |
response to pheromone(GO:0019236) |
| 0.0 |
0.8 |
GO:0000387 |
spliceosomal snRNP assembly(GO:0000387) |
| 0.0 |
0.1 |
GO:0072513 |
positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 |
0.1 |
GO:0030205 |
dermatan sulfate metabolic process(GO:0030205) |
| 0.0 |
0.3 |
GO:0002495 |
antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 |
0.5 |
GO:0006625 |
protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 |
0.6 |
GO:0000413 |
protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 |
0.1 |
GO:1901165 |
positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 |
0.1 |
GO:0000491 |
small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 |
0.1 |
GO:2000348 |
regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 |
0.3 |
GO:0045647 |
negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 |
0.1 |
GO:0090289 |
regulation of osteoclast proliferation(GO:0090289) |
| 0.0 |
0.0 |
GO:0018171 |
peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 |
0.3 |
GO:0070986 |
left/right axis specification(GO:0070986) |
| 0.0 |
0.1 |
GO:0043137 |
DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 |
0.1 |
GO:0006592 |
ornithine biosynthetic process(GO:0006592) |
| 0.0 |
0.6 |
GO:0007218 |
neuropeptide signaling pathway(GO:0007218) |
| 0.0 |
0.2 |
GO:0039702 |
viral budding via host ESCRT complex(GO:0039702) |
| 0.0 |
0.0 |
GO:0019405 |
alditol catabolic process(GO:0019405) |
| 0.0 |
0.1 |
GO:1903361 |
protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 |
0.6 |
GO:0007026 |
negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 |
0.1 |
GO:0048227 |
plasma membrane to endosome transport(GO:0048227) |
| 0.0 |
0.2 |
GO:0006122 |
mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 |
0.4 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 |
0.1 |
GO:0046085 |
adenosine metabolic process(GO:0046085) |
| 0.0 |
0.1 |
GO:0070127 |
tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 |
0.1 |
GO:0070234 |
positive regulation of T cell apoptotic process(GO:0070234) |
| 0.0 |
0.1 |
GO:0008206 |
bile acid metabolic process(GO:0008206) |
| 0.0 |
0.1 |
GO:0072531 |
pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.0 |
0.2 |
GO:0006183 |
GTP biosynthetic process(GO:0006183) |
| 0.0 |
0.2 |
GO:0051974 |
negative regulation of telomerase activity(GO:0051974) |
| 0.0 |
0.1 |
GO:0060068 |
vagina development(GO:0060068) |
| 0.0 |
0.1 |
GO:0060217 |
hemangioblast cell differentiation(GO:0060217) |
| 0.0 |
0.3 |
GO:0030213 |
hyaluronan biosynthetic process(GO:0030213) |
| 0.0 |
0.1 |
GO:0000291 |
nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.0 |
0.0 |
GO:0032962 |
regulation of inositol trisphosphate biosynthetic process(GO:0032960) positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 |
0.2 |
GO:0071260 |
cellular response to mechanical stimulus(GO:0071260) |
| 0.0 |
0.1 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.0 |
0.3 |
GO:0044342 |
type B pancreatic cell proliferation(GO:0044342) |
| 0.0 |
0.1 |
GO:0006049 |
UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 |
0.1 |
GO:0048262 |
determination of dorsal/ventral asymmetry(GO:0048262) determination of dorsal identity(GO:0048263) |
| 0.0 |
0.2 |
GO:0050819 |
negative regulation of coagulation(GO:0050819) |
| 0.0 |
0.0 |
GO:0046032 |
ADP catabolic process(GO:0046032) |
| 0.0 |
0.3 |
GO:1900452 |
regulation of long term synaptic depression(GO:1900452) |
| 0.0 |
0.1 |
GO:0018065 |
protein-cofactor linkage(GO:0018065) |
| 0.0 |
0.2 |
GO:0006012 |
galactose metabolic process(GO:0006012) |
| 0.0 |
0.3 |
GO:0070200 |
establishment of protein localization to telomere(GO:0070200) |
| 0.0 |
0.0 |
GO:0036388 |
pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 |
0.3 |
GO:0071470 |
cellular response to osmotic stress(GO:0071470) |
| 0.0 |
0.1 |
GO:0007076 |
mitotic chromosome condensation(GO:0007076) |
| 0.0 |
0.1 |
GO:0048669 |
collateral sprouting in absence of injury(GO:0048669) |
| 0.0 |
0.0 |
GO:0002578 |
negative regulation of antigen processing and presentation(GO:0002578) |
| 0.0 |
0.2 |
GO:0060716 |
labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 |
0.2 |
GO:2000177 |
regulation of neural precursor cell proliferation(GO:2000177) |
| 0.0 |
0.0 |
GO:0034143 |
regulation of toll-like receptor 4 signaling pathway(GO:0034143) |
| 0.0 |
0.0 |
GO:0000578 |
embryonic axis specification(GO:0000578) |
| 0.0 |
0.1 |
GO:0002667 |
lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.0 |
0.1 |
GO:1900028 |
negative regulation of ruffle assembly(GO:1900028) |
| 0.0 |
0.3 |
GO:1903077 |
negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 |
0.2 |
GO:0042832 |
defense response to protozoan(GO:0042832) |
| 0.0 |
0.1 |
GO:0032483 |
regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 |
0.2 |
GO:0022027 |
interkinetic nuclear migration(GO:0022027) |
| 0.0 |
0.0 |
GO:2000569 |
positive regulation of interleukin-17 production(GO:0032740) T-helper 2 cell activation(GO:0035712) positive regulation of T-helper 17 type immune response(GO:2000318) regulation of T-helper 2 cell activation(GO:2000569) positive regulation of T-helper 2 cell activation(GO:2000570) |
| 0.0 |
0.1 |
GO:0051204 |
protein insertion into mitochondrial membrane(GO:0051204) |
| 0.0 |
0.1 |
GO:0044539 |
long-chain fatty acid import(GO:0044539) |
| 0.0 |
0.1 |
GO:0046889 |
positive regulation of lipid biosynthetic process(GO:0046889) |
| 0.0 |
0.1 |
GO:0046598 |
positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 |
0.0 |
GO:1904154 |
positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 |
0.1 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.0 |
0.1 |
GO:0071028 |
nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 |
0.1 |
GO:1904152 |
negative regulation of protein exit from endoplasmic reticulum(GO:0070862) regulation of retrograde protein transport, ER to cytosol(GO:1904152) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 |
0.1 |
GO:0090043 |
regulation of tubulin deacetylation(GO:0090043) |
| 0.0 |
0.1 |
GO:2000051 |
negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 |
0.2 |
GO:0035855 |
megakaryocyte development(GO:0035855) |
| 0.0 |
0.3 |
GO:0031365 |
N-terminal protein amino acid modification(GO:0031365) |
| 0.0 |
0.7 |
GO:0050810 |
regulation of steroid biosynthetic process(GO:0050810) |
| 0.0 |
0.1 |
GO:0071803 |
positive regulation of podosome assembly(GO:0071803) |
| 0.0 |
0.2 |
GO:0060236 |
regulation of mitotic spindle organization(GO:0060236) |
| 0.0 |
0.0 |
GO:0006167 |
AMP biosynthetic process(GO:0006167) |
| 0.0 |
0.2 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 |
0.1 |
GO:1904707 |
regulation of vascular smooth muscle cell proliferation(GO:1904705) positive regulation of vascular smooth muscle cell proliferation(GO:1904707) vascular smooth muscle cell proliferation(GO:1990874) |
| 0.0 |
0.2 |
GO:0007614 |
short-term memory(GO:0007614) |
| 0.0 |
0.1 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 |
0.0 |
GO:0006788 |
heme oxidation(GO:0006788) |
| 0.0 |
0.0 |
GO:0042138 |
meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 |
0.1 |
GO:0060040 |
retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 |
0.1 |
GO:0032060 |
bleb assembly(GO:0032060) |
| 0.0 |
0.1 |
GO:0035025 |
positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 |
0.2 |
GO:0006677 |
glycosylceramide metabolic process(GO:0006677) |
| 0.0 |
0.1 |
GO:0006297 |
nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 |
0.0 |
GO:0035630 |
bone mineralization involved in bone maturation(GO:0035630) regulation of chondrocyte development(GO:0061181) regulation of bone mineralization involved in bone maturation(GO:1900157) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.0 |
0.1 |
GO:1903301 |
positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 |
0.1 |
GO:0001834 |
trophectodermal cell proliferation(GO:0001834) |
| 0.0 |
0.1 |
GO:0015868 |
purine ribonucleotide transport(GO:0015868) |
| 0.0 |
0.3 |
GO:0010972 |
negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 |
0.1 |
GO:0035721 |
intraciliary retrograde transport(GO:0035721) |
| 0.0 |
0.0 |
GO:1903300 |
negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 |
0.2 |
GO:0032981 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |